BACKGROUND
Molecular profiling is the screening of molecules to identify deoxyribonucleic acid (DNA), ribonucleic acid (RNA) and protein. Over the last 45-years, gene sequencing has become a key technology in many areas of biology and other sciences including medicine. The advent of rapid DNA, RNA and protein sequencing methods has greatly accelerated biological and medical research and discovery.1,2 Its application to medicine continues to help further elucidate the biological nature of disease processes and provides a better understanding of genetic diseases by allowing for an ‘up close’ look at the genomic signature or fingerprint of diseases.
The advent of technology, such as transcriptomics, has allowed for a better understanding and a more precise definition of disease.3,4 This has shifted focus to specific pathways and the genomic effects of disease processes. Transcriptomics has made it possible to better predict prognosis and tailor care providers’ approach to treatment.5,6,7 Molecular profiling has great potential. It has permitted targeted therapy to become the mainstay of standard of care, in many cases, resulting in statistical differences in patient morbidity and mortality.
Transcriptomics can predict lineage ‘trajectories’, however, the results do not necessarily reflect genetic relationships.
Therefore, techniques such as ‘lineage tracing’ are used to further describe single-cell genetic lineage. Lineage tracing uses a single cell marker to delineate all progeny produced by a single cell. The information provided is essential for understanding tissue heterogeneity within a given cell population and allows for greater interpretation of tissue disease and development.8
Sequencing individual genes, gene regions, or sets of genes, is becoming the leading approach to screening known or discovering novel germ lines and somatic mutations in focused areas of the genome.6,7 This article focuses on kidney disease and describes how our understanding of kidney disease pathogenesis has been limited by an incomplete molecular characterization of the cell types responsible for the organ’s multiple homeostatic functions.9,10
This review was initiated in an attempt to identify, characterize, and discuss literature supporting clinically relevant molecular markers and interventions. The efficacy of targeting specific markers will be examined with data from clinical trials focusing on treatments for different disease process. Herein, we review the application of single-cell transcriptomics on mouse kidneys and briefly describe how its application has the potential to elucidate and provide a better understanding of cellular function and disease patterns.
Identification of a Novel Cell Type Using Fluorescent Staining and Lineage Tracing
There are at least two distinct tubule epithelial cells within the collecting duct of the kidney, the principle cells (PC) and intercalated cells (IC). These cells play important roles in fluid and electrolyte balance and acid-base homeostasis respectively.11 ICs can be further subdivided to alpha and beta-ICs. Alpha-ICs have an apical H+ ATPase and basolateral chloride-bicarbonate exchanger; whereas beta-ICs have a basolateral proton ATPase and apical chloride-anion exchanger pendrin.
Studies have suggested that certain epithelial cells such as beta-ICs are not static. Rather, certain factors that alter cellular biology are known to remodel cells as a way of adapting to stressors. Schwartz et al demonstrated that changes in acid-base homeostasisin particular metabolic acidosis can convert beta-ICs to alphaICs. The other researchers have evaluated cellular plasticity with hopes of determining whether a common intermediate epithelium element exists.12
A newly defined cell type, described as transitional cells, was characterized by Park et al.9 They discovered that these cells expressed key marker genes that define PC (AQP2, Aquaporin 2) and IC cells (ATP6V1G3, H+-ATPase) in addition to cell typespecific markers such as Parm1 and Section 23b (Figure 1).
Figure 1. Heatmap Showing the Expression Level of Differentially Expressed Genes in Collecting Duct Cell Types. Color scheme is Based on Z-score Distribution.
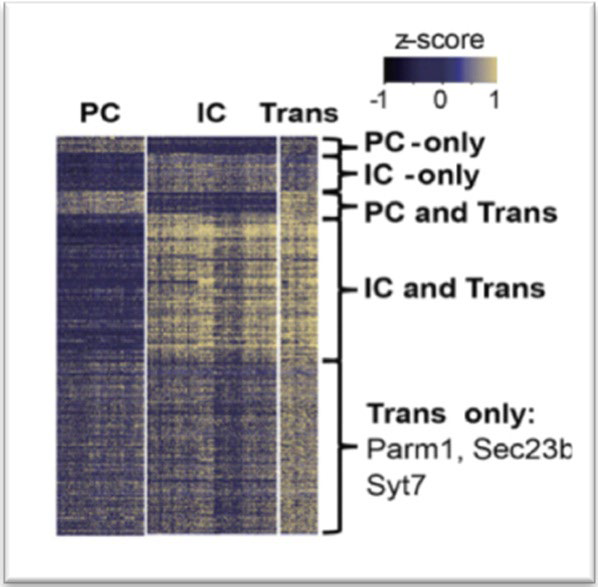
They confirmed their existence through double immunofluorescence staining and in situ hybridization using gene specific probes in mice. The expression of low stress response and cell cycle genes were monitored to ensure that the newly discovered cells were not ‘miss labeled’ as proliferating progenitor cells, injured cells or artifact. Follow-up with in vivo lineage tracing experiments was used to confirm that intercalated cells and principal cells have the ability to interconvert by means of the newly identified transitional cell type. This discovery prompted further investigation into the transitional cell type, specifically their plasticity and the influencers of cellular transition.
Cellular Plasticity as a Result of Notch Ligands and Receptors
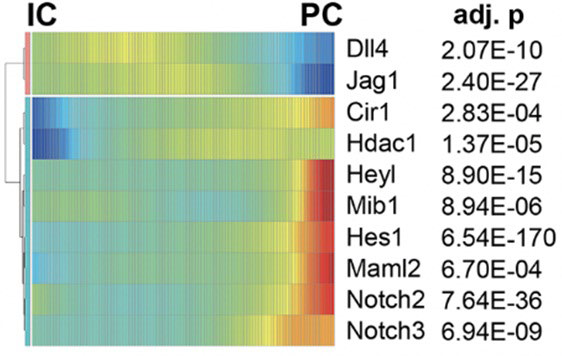
To further investigate collecting duct cell plasticity, the researchers identified genes with variable expression levels during cellular transitions. Gene expression patterns suggested that Notch signaling pathway was activated during the transition from intercalated cells to principal cells (Figure 2). They devised an experimental model to study the IC to PC transition using an inducible transgenic mouse. Their results demonstrated that Notch signaling and receptor expression is sufficient to drive cell transition in differentiated adult kidney collecting tubule. Furthermore, they examined whether there is increased Notch expression in kidney disease states by analyzing the relative numbers of PC and IC cells after inducing chronic kidney disease in a mouse model. Computational and gene expression analysis was consistent with an increased shift of IC to PC cells in the diseased tissue samples. Furthermore, the increased IC to PC transition was found to be a likely cause of metabolic acidosis in diseased kidney states, a consequence from the shift toward the PC cell fate.
Figure 2. Transcriptional Profiles Demonstrating the Spectrum of Expression of Notch Genes in the Collecting Duct. Cells are Ordered in Pseudotime and Color Represents Expression Levels
DISCUSSION
The research performed by Park et al, complements previous work where single-cell RNA sequencing was used in mice to identify transcriptomes for the major collecting duct cell types.9 In contrast to identifying a novel cell type, Chen et al demonstrated considerable heterogeneity within each of the three cell types in the kidney collecting tubule. They speculated that transcription of cell type-specific markers is not a static event and therefore messenger ribonucleic acid (mRNA) coding in individual cell types may have varying degrees of expressivity.13,14 However, similarly, they found cells that express transcripts known to be specific to other cell types. At the time of their study, they classified these cells as hybrid IC/PCs.15 Therefore, it is possible that their discovery of ‘hybrid cells’ is consistent with the ‘transitional cell’ described by Park et al. The data gathered allows for the identification of genes that are expressed selectively in different cell types. The data from studies experimenting with single-cell RNA sequencing is useful for a variety of applications including potential drug targets, understanding of kidney physiology, kidney disease pathogenesis, and tracing genetic disease of the kidney to a single cell type.
The plasticity of the renal epithelial cells has been explored in renal healing mechanisms in response to acute kidney injury (AKI).14,16 As previously mentioned, Park et al demonstrated that diseased tissue samples showed a higher ratio of PC to IC cells as a result of variable gene expression.9 In a study by Trepiccione et al12 the cellular plasticity in the collecting duct during recovery from lithium-induced injury was examined. They too identified a novel cell type positive for both IC and PC cell markers on average six days after lithium washout. They suggest that these cells drive the reversal of cellular change seen during the recovery period of Li+-induced nephrogenic diabetes insipidus and could represent a transition element in the conversion of IC’s into PC’s.12 The physiological role and developmental significance of these new cells remains to be fully determined and may be the subject of future research.
CONCLUSION
Molecular profiling and precision therapy are revolutionizing the field of medicine. Single-cell transcriptomics of the mouse kidney revealed potential cellular targets of kidney disease. The application of sequencing technology led to the identification of cell-type specific markers along with novel cell types and led to the uncovering of unexpected cell plasticity.4 This targeted approach has potential for many benefits. The identification of novel cell lines allows for a more accurate understanding of kidney physiology and the diagnosis of kidney disease and precise staging of disease.17,18 Through the use of technologies such as immunohistochemical analysis, long-standing biophysical questions have been answered by the discovery of protein complexes.19-22 Park et al utilized computational cell trajectory analysis and in vivo lineage tracing and revealed that intercalated cells and principal cells undergo transitions mediated by the Notch signaling pathway, which were shifted toward a principal cell fate and were associated with metabolic acidosis.9
The continued application of resources and research in understanding the biochemical environment and cellular mechanisms has allowed for a more thorough understanding of normal kidney function and disease development. Molecular profiling and precision therapy have the potential for enhancement in effective therapy for a precise subset of patients, whom stand to benefit the most.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.







