INTRODUCTION
In Mexico, most of the population (>90%) is the consequence of an admixture on account of European contact principally with the Spaniards, Amerindians, and African slaves. This part of the population is commonly referred to as the Mestizos, who speak Spanish and live in both urban and rural regions throughout the country. Many studies have been conducted in Mexican-Mestizos to validate the application of autosomal short tandem repeats (STRs) for human identification purposes.1,2 However, in Mexico, this forensic validation requires the study of different populations, given the genetic structure demonstrated among Mestizos from different regions.3 Based on the 13 STRs of the Combined DNA Index System (CODIS), it has been claimed that the European ancestry increases gradually towards the Northwestern region and diminishes towards the Central and Southeastern regions; conversely, the Amerindian ancestry displays an opposite pattern increasing towards the Central and Southeastern region, and decreasing towards the Northwestern region.1,2 Although, several Mexican-Mestizo populations have been studied, some geographical areas have been poorly analyzed for STR markers from a forensic perspective; particularly the Southern region characterized by the presence of numerous Mexican indigenous communities, which could potentially increase the Amerindian ancestry of the neighboring southern Mestizo populations.4 Therefore, the two defined objectives of this study were:
1. To report the statistical parameters of forensic efficiency for 15 STRs of the Identifiler® kit in the state of Guerrero, a representative of the southern Mestizo population;
2. To compare the collected data with the populations from the main geographical regions of Mexico.
MATERIALS AND METHODS
DNA Samples
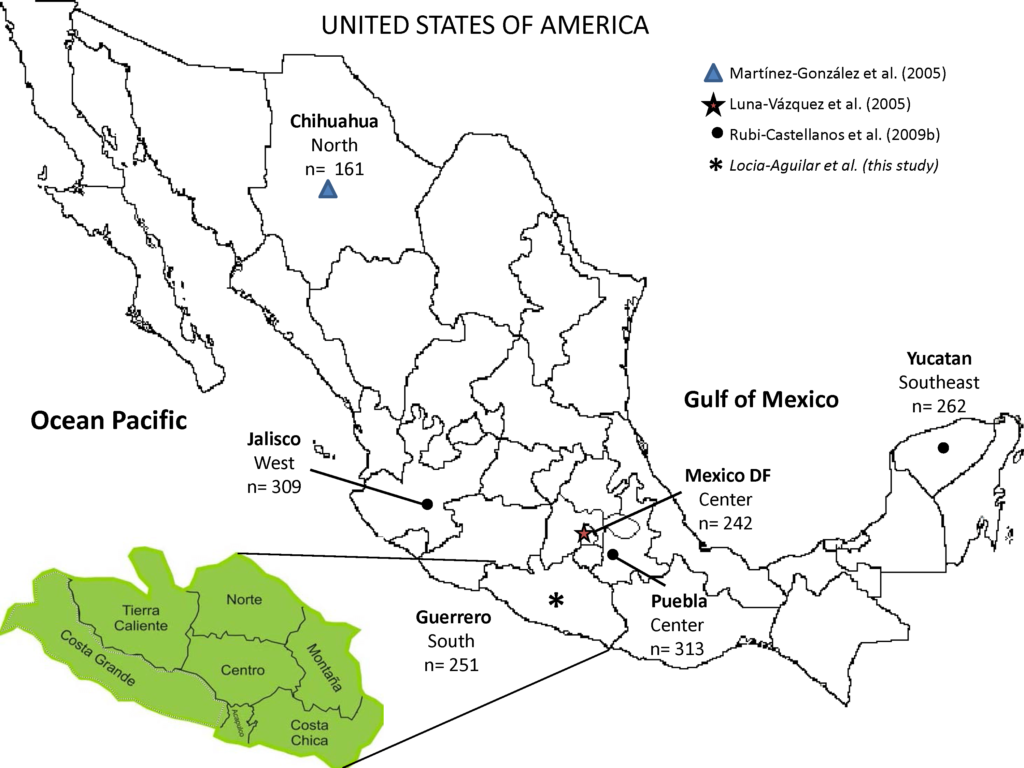
We analyzed the DNA samples of 251 unrelated Mexican-Mestizo individuals from the following regions of the southern state of Guerrero: Costa Chica (n=14), Acapulco (n=80), Centro (n=53), Montaña (n=23), Norte (n=56), and Tierra Caliente (n=25) (Figure 1). All individuals participating in the study, signed a written informed consent according to the ethical guidelines of the Helsinki Declaration.
STR Genotyping
DNA was extracted from a dried blood specimen spotted on the FTA paper or buccal swabs using Chelex® 100 or the standard phenol-chloroform method. Genotypes for 15 STRs were obtained with the AmpFlSTR Identifiler® kit followed by capillary electrophoresis according to the supplier´s instructions using the ABI-Prism 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Allele ladder provided in the kit and the software Genemapper® 3.2 were used for allele calling.
Quality Control
Positive and negative controls were used as specified in the Identifiler® kit user’s manual. The genotype data was analyzed and verified independently by two different statistical methods.
Data Analysis
Allele frequencies and statistical parameters of forensic efficiency were estimated with the spreadsheet PowerStats.5 Hardy-Weinberg equilibrium (HWE) for each and the combined loci were computed by performing the exact tests using the software Genetic Data Analysis (GDA) 1.1.6 Population differentiation was evaluated by performing pairwise comparisons (exact test p-value) and normalized FST genetic distances represented in a Multidimensional scaling (MDS) plot using the software Arlequin 3.07 and Statistical Package for the Social Sciences (SPSS) version 20.0, respectively. For this purpose, four previously reported Mexican-Mestizo populations from different geographic regions were included in the interpopulational analysis: Chihuahua (North),8 Jalisco (West),9 Valley of Mexico,10 and state of Puebla (Center),9 and Yucatan (Southeast)9 (Figure 1).
Figure 1: Geographical Location, Sample Size (n) and Reference of the Mexican-Mestizo Populations Used for Interpopulation Analysis in this Study
RESULTS AND DISCUSSIONS
Allele frequencies and statistical parameters of forensic efficiency of the 15 STRs for the Identifiler® kit have been summarized in Table 1. In addition, complete genotype STR dataset is available as electronic supplementary material in Supplementary Table 1. This information will be helpful to the Mexican forensic geneticists to evaluate paternity and forensic cases solved in the Guerrero state. According to these forensic parameters, the most informative loci were D18S51, D2S1338 and FGA; conversely, the STRs with the lowest forensic informativity were TPOX and D3S1358. The combined power of exclusion for the STR genetic system was 99.999634444%, whereas the combined power of discrimination was greater than >99.99999999%, confirming the potential of this kit for human identification purposes. Although, HWE test showed a low p-value for D21S11 (p=0.041), the application of the Bonferroni correction (p<0.0033) suggested that the genotype distribution was in agreement with the Hardy-Weinberg expectations for all the 15 STR loci in the population of Guerrero. Similarly, the exact linkage equilibrium was established between all pairs of loci (p>0.0038) (Data not shown). In brief, these results suggested that the binomial square and product rule can be confidently used in forensic caseworks to estimate DNA profiles obtained with the Identifiler® kit in the population of Guerrero.
| Table 1. Allele Frequency Distribution and Statistical Parameters of Forensic Efficiency of 15 STRs (Identifiler kit) in 251 Mexican-Mestizos from the State of Guerrero (South, Mexico). |
|
Allele
STR
|
D8S1179 |
D21S11 |
D7S820 |
CSF1PO |
D3S1358 |
THO1 |
D13S317 |
D16S539 |
D2S1338 |
D19S433 |
VWA |
TPOX |
D18S51 |
D5S818 |
FGA |
| 2 |
|
|
|
0.0020 |
|
|
|
|
|
|
|
0.0020 |
|
|
|
| 4 |
|
|
|
|
|
0.0020 |
|
|
|
|
|
|
|
|
|
| 6 |
|
|
|
0.0020 |
|
0.3406 |
|
|
|
|
|
0.0020 |
|
|
|
| 7 |
|
|
0.0060 |
0.0040 |
|
0.3845 |
|
|
|
|
|
|
|
0.0797 |
|
| 8 |
|
|
0.0697 |
0.0080 |
|
0.0378 |
0.0600 |
0.0060 |
|
|
|
0.4741 |
|
0.0080 |
|
| 9 |
|
|
0.0299 |
0.0259 |
|
0.0598 |
0.2520 |
0.1195 |
|
|
|
0.0359 |
|
0.0618 |
|
| 9.3 |
|
|
|
|
|
0.1713 |
|
|
|
|
|
|
|
|
|
| 10 |
0.0996 |
|
0.2550 |
0.2390 |
|
0.0040 |
0.1240 |
0.1932 |
|
0.0020 |
|
0.0378 |
0.0020 |
0.0538 |
|
| 11 |
0.0518 |
|
0.3446 |
0.2610 |
|
|
0.1560 |
0.2928 |
|
|
|
0.2769 |
0.0120 |
0.4801 |
|
| 12 |
0.1315 |
|
0.2351 |
0.3884 |
|
|
0.2120 |
0.3028 |
|
0.0378 |
|
0.1614 |
0.0777 |
0.2371 |
|
| 12.2 |
|
|
|
|
|
|
|
|
|
0.0139 |
|
|
|
|
|
| 13 |
0.3805 |
|
0.0478 |
0.0637 |
0.0080 |
|
0.1140 |
0.0777 |
|
0.1793 |
0.0020 |
0.0100 |
0.1633 |
0.0737 |
|
| 13.2 |
|
|
|
|
|
|
|
|
|
0.1255 |
|
|
|
|
|
| 14 |
0.2311 |
|
0.0120 |
0.0040 |
0.0498 |
|
0.0820 |
0.0080 |
|
0.2530 |
0.0598 |
|
0.1693 |
0.0060 |
|
| 14.2 |
|
|
|
|
|
|
|
|
|
0.0717 |
|
|
|
|
|
| 15 |
0.0837 |
|
|
0.0020 |
0.5060 |
|
|
|
|
0.1096 |
0.0837 |
|
0.1394 |
|
|
| 15.2 |
|
|
|
|
|
|
|
|
|
0.1295 |
|
|
|
|
|
| 16 |
0.0159 |
|
|
|
0.2649 |
|
|
|
0.0179 |
0.0458 |
0.3805 |
|
0.0976 |
|
|
| 16.2 |
|
|
|
|
|
|
|
|
|
0.0239 |
|
|
|
|
|
| 17 |
0.0040 |
|
|
|
0.1016 |
|
|
|
0.1295 |
0.0020 |
0.2590 |
|
0.1773 |
|
|
| 17.2 |
|
|
|
|
|
|
|
|
|
0.0020 |
|
|
|
|
|
| 18 |
0.0020 |
|
|
|
0.0657 |
|
|
|
0.0598 |
0.0040 |
0.1494 |
|
0.0717 |
|
0.0060 |
| 18.2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.0020 |
| 19 |
|
|
|
|
0.0020 |
|
|
|
0.2331 |
|
0.0458 |
|
0.0359 |
|
0.0797 |
| 20 |
|
0.0020 |
|
|
0.0020 |
|
|
|
0.1773 |
|
0.0179 |
|
0.0239 |
|
0.0378 |
| 21 |
|
|
|
|
|
|
|
|
0.0159 |
|
0.0020 |
|
0.0139 |
|
0.1215 |
| 22 |
|
|
|
|
|
|
|
|
0.0618 |
|
|
|
0.0080 |
|
0.0936 |
| 23 |
|
|
|
|
|
|
|
|
0.2151 |
|
|
|
0.0040 |
|
0.1414 |
| 23.2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.0020 |
| 24 |
|
|
|
|
|
|
|
|
0.0657 |
|
|
|
0.0040 |
|
0.1514 |
| 24.2 |
|
0.0020 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 25 |
|
|
|
|
|
|
|
|
0.0239 |
|
|
|
|
|
0.1972 |
| 25.2 |
|
0.0020 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 26 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.1056 |
| 27 |
|
0.0100 |
|
|
|
|
|
|
|
|
|
|
|
|
0.0458 |
| 28 |
|
0.0876 |
|
|
|
|
|
|
|
|
|
|
|
|
0.0120 |
| 29 |
|
0.1912 |
|
|
|
|
|
|
|
|
|
|
|
|
0.0020 |
| 30 |
|
0.2629 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 30.2 |
|
0.0100 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 31 |
|
0.1096 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 31.2 |
|
0.1155 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 32 |
|
0.0199 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 32.2 |
|
0.1215 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 33.2 |
|
0.0578 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 34.2 |
|
0.0040 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 35.2 |
|
0.0020 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 36 |
|
0.0020 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 43.2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0.0020 |
| MAF |
0.0114 |
0.0120 |
0.0112 |
0.0114 |
0.0106 |
0.0107 |
0.0126 |
0.0116 |
0.0127 |
0.0119 |
0.0111 |
0.0108 |
0.0123 |
0.0108 |
0.0124 |
| PD |
0.9113 |
0.9537 |
0.8994 |
0.8615 |
0.8389 |
0.8540 |
0.9460 |
0.9021 |
0.9500 |
0.9582 |
0.9039 |
0.8344 |
0.9687 |
0.8572 |
0.9687 |
| PE |
0.5218 |
0.6380 |
0.4681 |
0.5218 |
0.3490 |
0.3654 |
0.7306 |
0.5498 |
0.7557 |
0.6229 |
0.4553 |
0.3882 |
0.6921 |
0.3999 |
0.7079 |
| Het |
0.7570 |
0.8207 |
0.7251 |
0.7570 |
0.6454 |
0.6574 |
0.8680 |
0.7729 |
0.8805 |
0.8127 |
0.7171 |
0.6733 |
0.8486 |
0.6813 |
0.8566 |
| PIC |
0.7336 |
0.8244 |
0.7130 |
0.6708 |
0.6093 |
0.6490 |
0.8062 |
0.7269 |
0.8186 |
0.8335 |
0.7178 |
0.6177 |
0.8571 |
0.6579 |
0.8605 |
| TPI |
2.0574 |
2.7889 |
1.8188 |
2.0574 |
1.4101 |
1.4593 |
3.7879 |
2.2018 |
4.1833 |
2.6702 |
1.7676 |
1.5305 |
3.3026 |
1.5688 |
3.4861 |
| HWE |
0.2509 |
0.0041 |
0.3088 |
0.4884 |
0.5088 |
0.0131 |
0.2281 |
0.7966 |
0.4156 |
0.6506 |
0.4413 |
0.0369 |
0.8978 |
0.1591 |
0.2472 |
| MAF: Minimum allele frequency; HWE: p-value of exact tests for Hardy-Weinberg equilibrium expectations; PD: Power of Discrimination; PE. Power of Exclusion; Het. Heterozygosity; PIC. Polymorphism Informativity Content; TPI: Typical Paternity Index. |
The knowledge of the genetic relationships among populations is important because it guides the work of forensic geneticists to analyze alternative STR population data during the statistical interpretation, given that most of the Mexican-Mestizo populations do not have their own STR database. Until now, this interpopulation analyses of Mestizo populations including the main geographic regions of the Mexican territory have been carried out only with the 13 CODIS-STRs.1,2 Therefore, we represented the genetic distances among six Mexican-Mestizo populations in an multidimensional scaling (MDS) plot with the complete set of 15 STRs constituting the Identifiler kit (Figure 2). Although, recently, three Mexican Native populations from Guerrero were analyzed with these 15 STRs,11 the complete genotype dataset was not available and thus, could not be included in this report. The close similarity observed between the southern state of Guerrero and the central populations is partially in agreement with the geographical parameters. However, the greater extent of similarity between the population residing in the Valley of Mexico with Guerrero (p=0.2342) than with the nearby population of Puebla (p=0.0000), disregards the idea that geography could possibly explain such genetic associations (Table 2). This observation is in agreement with the first global study conducted on Mexican-Mestizos based on CODIS-STRs that discarded the Isolation by Distance (IBD) hypothesis1 by means of the autocorrelation indices for DNA analysis (AIDA). These contradictory results suggest that Mexican population relationships have been explained by factors such as socioeconomic conditions and educational status associated with the higher European ancestry.12 Similarly, the IBD hypothesis has not been followed for the study of Native American populations from Mexico when geographic distances are lesser than 1500 km,13 which is in agreement with our results. A different quadrant of the MDS plot represents the populations of Chihuahua (North) and Jalisco (West), whereas the Yucatan population is isolated in a third different section of the plot (Figure 2). Although, this result is partially in agreement with a previous description that roughly classifies the distribution of the Mexican-Mestizo population into two principal regions (Northwest and Center-Southeast).1,2 The observed distribution suggests that the Southeastern region is different and should be considered as independent from the Central and Southern regions.
Figure 2: MDS Plot Representing the Genetic Relationships Among Six Mexican-Mestizo Populations Based upon the Powerplex 16 System.
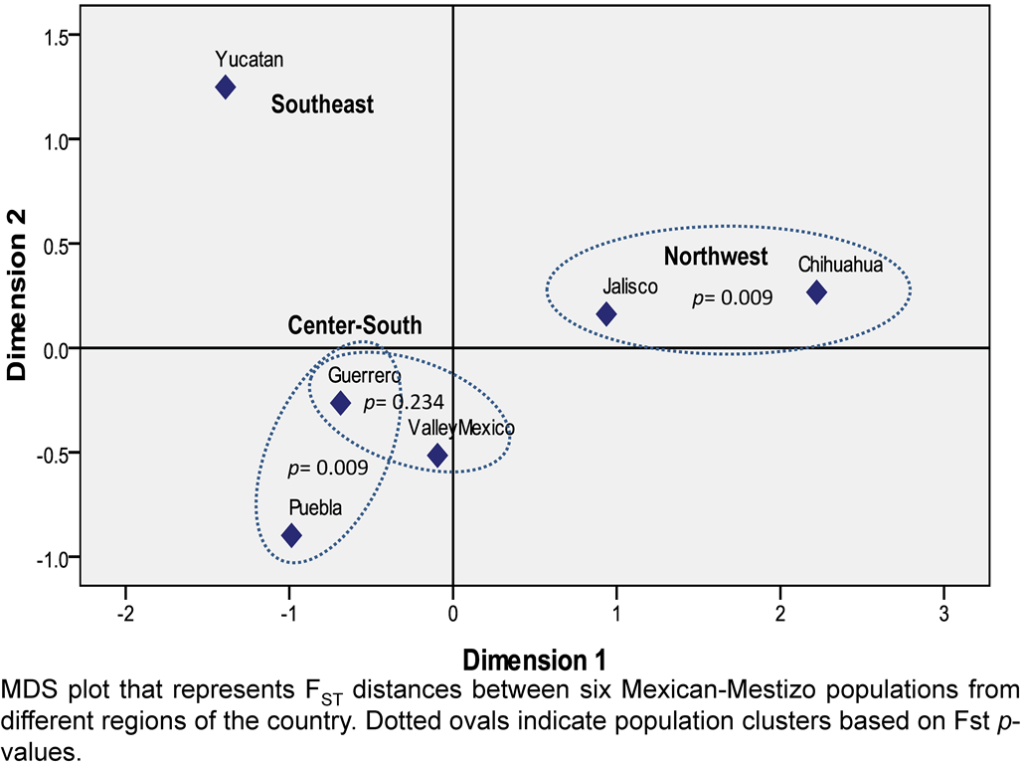
| Table 2: Pairwise FST p-values (above diagonal) and FST Genetic Distances (below diagonal) between Five Mexican-Mestizo Populations from Different Geographic Regions based on 15 STR Loci. |
|
Chihuahua
North |
Jalisco
West |
Valley Mexico
Center |
Puebla
Center |
Guerrero
South |
Yucatan
Southeast |
| Chihuahua |
**** |
0.00901 |
0.00000 |
0.00000 |
0.00000 |
0.00000 |
| Jalisco |
0.00118 |
**** |
0.00000 |
0.00000 |
0.00000 |
0.00000 |
| Valley Mexico |
0.00484 |
0.00125 |
**** |
0.00000 |
0.23423 |
0.00000 |
| Puebla |
0.00928 |
0.00368 |
0.00108 |
**** |
0.00901 |
0.00000 |
| Guerrero |
0.00777 |
0.00264 |
0.00028 |
0.00104 |
**** |
0.00000 |
| Yucatan |
0.01029 |
0.00559 |
0.00409 |
0.00419 |
0.0027 |
**** |
CONCLUSION
We estimated for the first time the forensic parameters for 15 STRs in Mestizos from Guerrero (South, Mexico). We describe the genetic structure of Mexican-Mestizos based on 15 STRs, where the Southeastern region showed differences with the Central and Southern regions.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.







