INTRODUCTION
Lung cancer remains the leading cause of cancer-related death worldwide, accounting for 1.59 million of the 8.2 million total cancer deaths each year.1 NSCLC accounts for ~80% of all lung cancers, and adenocarcinoma is the main histological subtype. Smoking is responsible for 87% of lung cancers in the United States, and the majority of NSCLC adenocarcinomas. While historically, cigarette smoking was associated with NSCLC squamous cell lung cancer, since the transition to filtered cigarettes there has been a rise in the number of adeocarcinomas among smokers. Most NSCLC patients are diagnosed at an advanced stage that is associated with high recurrence and less than 15% survival over 5 years. Even for patients with Stage I disease who have had a complete surgical resection, the 5-year survival is only 52%,2 mostly because of recurrence. This suggests that Stage I tumors can be misclassified, especially when lymph node metastases are small and escape detection. Furthermore, intratumor heterogeneity may not be detected by standard singlesite biopsy, leading to inaccurate classification and prognosis.3,4 Classification and staging by Tumor, Node, Metastasis (TNM)5 have only modest prognostic utility and patients with resected Stage IA tumors of identical histology, differentiation, vascular invasion, and margins may differ widely in their survival time and response to therapy. The use of alternative sources of biopsy material that reflect tumor heterogeneity, such as readily available blood or plasma, may provide valuable information.
Cells and cellular nucleic acids in sputum, bronchial biopsies, brushing specimens, bronchial lavage fluid, blood, pleural effusions and solid tumor biopsies provide material for classification, diagnosis and prognosis of NSCLC.6 Several cellular tumor markers have been shown to be useful in prognosis of lung cancer including positive immunohistochemical staining of mTOR in early stage NSCLC,7 overexpression of Apolipoprotein E in lung adenocarcinomas with malignant pleural effusion,8 and overexpression of SOX2 in Stage I adenocarcinomas, especially those with pleural invasion.9 Recently, a 14-gene expression assay has been developed to examine recurrence risk in early-stage NSCLC.10
Small non-coding RNAs such as snoRNAs11 and microRNAs (“oncomirs”)12 are a promising class of cancer biomarkers. The expression profiles of these highly stable molecules in tumor tissue and/or plasma have been associated with occurrence of NSCLC, tumor tissue type, stage of differentiation, and prognosis (for review see13). microRNAs are generally down-regulated in tumor tissue compared to matched normal tissue. However, tumor-derived microRNAs are found within exosomes or in free circulation in the plasma of cancer patients14 and have been shown to be generally up-regulated compared to plasma from normal donors.15 A panel of four plasma microRNAs (mir-486, -30d, -1 and -499) was recently shown to discriminate between NSCLC patients with good and poor prognosis.15Serum levels of miR-142-3p and miR-29b were elevated in lung adenocarcinoma patients suffering recurrence within 24 months.16
There is an urgent need for better understanding of the molecular alterations that occur during progression of earlystage lung adenocarcinoma, and more reliable prognostic and predictive biomarkers. Such biomarkers could improve survival by identifying patients at high risk for recurrence who may benefit from adjuvant chemotherapy. Here we report a pilot study that determines microRNA profiles in tumors and plasma from patients with Stage I and Stage II/III adenocarcinoma, the most common form of NSCLC, and relate these findings to the biological processes that may be involved in early recurrence.
MATERIALS AND METHODS
Study population
Patient biopsy samples (frozen tumor and plasma) from Stage I (with no lymph node involvement) and Stage II/III (with lymph node involvement) were selected from the Lung Cancer Tumor Bank at Capital District Health Authority (CDHA; Halifax, NS, Canada). In order to limit the clinical variability as much as possible, a uniform study population was used based on the following criteria: adenocarcinoma, age >45 years; all but one were current or past smokers (Table 1).
Table 1: Patient characteristics and samples used for microRNA analysis.
|
ID
|
Stage
|
Age |
Gender |
Smoking Status |
Recurrence (months)
|
|
L202
|
IB |
76 |
Male |
Current |
|
| L218 |
IA |
65 |
Male |
Past
|
|
|
L247
|
IB |
77 |
Male |
Past |
|
| L252 |
IA |
81 |
Male |
Past
|
|
|
L272
|
IA |
79 |
Female |
Past |
|
| L278 |
IA |
69 |
Male |
Past
|
|
|
|
|
|
|
|
|
L194
|
IIA |
53 |
Male |
Past |
12.8 |
| L212 |
IIA |
71 |
Male |
Current |
|
|
L229
|
IIA |
66 |
Male |
Past |
8.9 |
| L240 |
IIA |
74 |
Male |
Past
|
|
|
L258
|
IIIA |
45 |
Male |
Never |
|
| L262 |
IIA |
76 |
Female |
Past
|
|
|
L300a
|
IIB |
54 |
Female |
Past
|
|
aremoved from analyses due to high hemolysis in plasma samples
Most cases of adenocarcinoma are associated with smoking, and these were highly represented in the Tumor Bank. Recurrence is the time in months from surgery until detection of recurrence. Control plasma (Precision Biologics, Dartmouth, NS, Canada) contained pooled plasma from 20 healthy subjects. This study was approved by the Capital Health Research Ethics Board (CDHA-RS/2004-336), and all participating individuals signed informed consent.
RNA extraction
Approximately 50 mg of frozen lung tissue was pulverized in a MultiSample BioPulverizer (BioSpec, Bartlesville, OK, USA) and homogenized in 1 mL TRIZOL® (Life Technologies, Burlington, ON, Canada) using a FastPrep®-24 (MP Biomedicals, Santa Ana, CA, USA). Total RNA was extracted according to the manufacturer’s protocol and treated with TURBO DNase (Applied Biosystems, Carlsbad, CA, USA) prior to further purification with the Total RNA Purification Kit (Norgen Biotech Corp., Thorold, ON, Canada). RNA was purified from 300 μL of plasma using the Total RNA Purification Kit and both preparations were enriched for small RNAs using the PureLink miRNA Isolation Kit (Invitrogen, Burlington, ON, Canada). The RNA was quantified on the NanoDrop-1000 (NanoDrop Products, Wilmington, DE, USA) and RNA quality was determined on the Bioanalyzer-2100 (Agilent, Wilmington, DE, USA).
MicroRNA assay of tissue and plasma
The miRNA PCR Array for Cancer (MAH-102F; SA Biosciences, Burlington, ON, Canada) containing 88 cancerrelevant microRNA-probes and 8 controls in a 96-well plate was employed to assess expression of microRNAs in pooled plasma samples of five Stage I (L202, 218, 247, 252 and 278) and five Stage II (194, 212, 229, 240 and 262) lung adenocarcinoma patients compared to pooled normal plasma (Precision Biologics). RNA was purified from 300 μL of plasma (60 μL each of five individual samples from each stage). 200 ng enriched small RNA was reverse-transcribed using the RT² First Strand Kit (SA Biosciences, Burlington, ON, Canada) and qPCR was performed on a Light Cycler-480 (Roche Applied Science, Laval, QC, Canada) using conditions specified by the manufacturer. Fold regulation was calculated from 2nd derivative Cp values using the SA Biosciences web-based qPCR analysis software (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.phphttp://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php) and SNORD48 was used as a reference.
Based on the results of the PCR array (Supp Table 1) and literature survey, a multiplex TaqMan® microRNA Assay (Applied Biosystems) was designed for 31 microRNAs that were at least two-fold up-regulated in cancer relative to control (Supp Table 2). RNU48 was used as a reference, allowing all 32 reactions to be performed in triplicate in 96-well plates. Expression was evaluated in an expanded group of six Stage I and seven Stage II/III tissue and plasma samples. Briefly, 100 ng of total RNA was reverse-transcribed using the TaqMan® microRNA Reverse Transcription Kit (Applied Biosystems) and qPCR reactions were cycled on a LightCycler-480 once at 95 °C for 10 min; 45 times at 95 °C for 15 sec, 60 °C for 60 sec; and once at 40 °C for 30 sec. Individual samples contained 1 μL cDNA, 5 μL of 2X qPCR mix and 1 μL of microRNA-specific TaqMan Probe/Primer mix in a 10 μL total reaction volume. For tumor samples, matched normal tissue samples in triplicate were used as controls and for plasma, pooled normal samples were used. In the latter case, control samples were assayed twice independently in triplicate reactions and the average was used.
Normalized data was analyzed by Student’s t-test and p-values were based on replicate 2nd derivative Cp values for each microRNA in the control group and patient group. Those microRNAs that were associated with high p-values (>0.05) were assigned as missing values. The Student’s t-test and SAM modules of MeV17 were then used to identify microRNAs that were significantly differentially expressed between the Stage I and Stage II/III samples. MicroRNA expression was analyzed from 2nd derivative Cp values using SA Bioscience web-based PCR data analysis software (Qiagen, Mississauga, ON, Canada) and the stably expressed RNU48 and miR-210 to normalize for tissue and plasma samples, respectively. Targets for microRNAs were identified by searching miRDB(www.mirDB.org). The list of microRNA targets with a score greater than 80 was used to identify KEGG pathways potentially involved in progression using gene enrichment analysis in WebGestalt (http://bioinfo. vanderbilt.edu/webgestalt).
RESULTS
In order to identify clinically relevant microRNA biomarkers, we screened 88 cancer-relevant microRNAs by array. We selected those that were up-regulated at least two-fold in plasma of lung adenocarcinoma patients relative to plasma from pooled normal subjects as these would be most easily detectable in a clinical assay. Sixty-eight microRNAs were up-regulated in plasma of NSCLC patients relative to normal subjects. Of these 44 microRNAs showed a higher level in Stage II/III vs. Stage I, 17 showed a lower level in Stage II/III vs. Stage I, and 7 did not differ between Stage II/III and Stage I (Table 2).
Of these, 20 for which there was corroborating evidence in the literature were chosen for TaqMan assays along with 11 other well-supported microRNAs from the literature (miR-17- 3p, 34a, 92a, 106a, 141, 182, 201, 218, 221, 451 and 486-5p; Supp Table 2).
Expression data for these 31 microRNAs were obtained using TaqMan assays for all tissue/matched normal samples and all but one plasma sample (L300 plasma was unusable due to ahigh level of lysed erythrocytes and was excluded from analysis). Several microRNAs (miR-17-3p, 141, 143, 205 and 218) exhibited inconsistent, often low, signal intensities in most plasma samples (grey boxes, Figure 1).
Figure 1: Heat map of miRNA expression in tumor tissue.
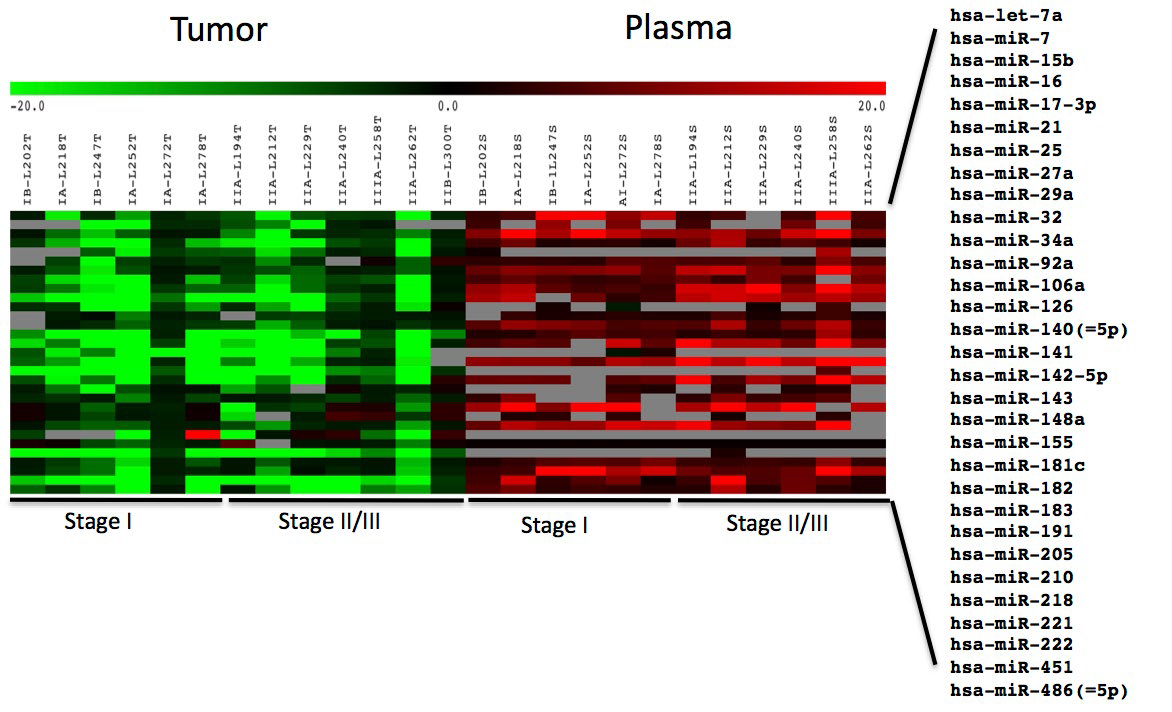
Expression of 31 miRNAs in tumor relative to matched normal tissue and patient plasma relative to pooled control plasma from six patients each with Stage I and Stage II/III NSCLC adenocarcinoma. Red, up-regulated; green, down-regulated; grey, high p-values (>0.05) were assigned as missing data. Log2 ratios are presented.
microRNAs were mostly down-regulated in tumor samples relative to normal adjacent tissue but up-regulated in plasma relative to control pooled plasma from normal subjects (Figure 1, Supp Table 3) . The single Stage IIIA sample (L258) exhibited generally higher, but not statistically significant, upregulation of plasma microRNAs than the other samples.
There were no statistically significant differences in tumor tissue microRNA expression between different stages.
However, SAM analysis identified seven plasma microRNAs that could distinguish between patients with Stage I and Stage II/III lung adenocarcinoma (Figure 2).
Figure 2: Heat map of miRNA expression in plasma.
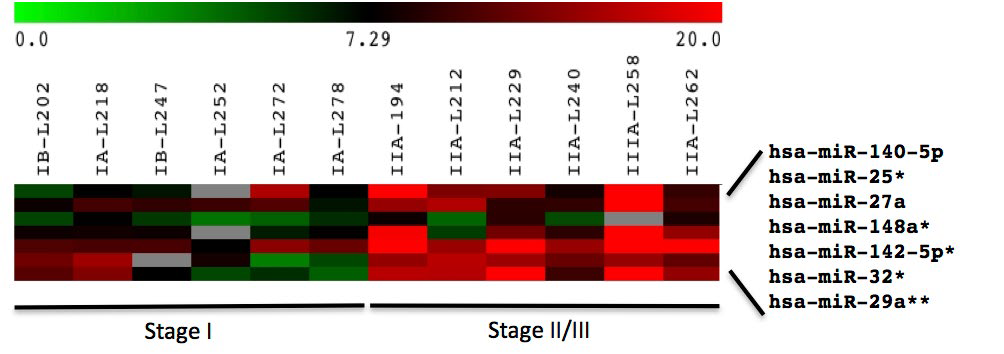
Expression of 31 miRNAs in tumor relative to matched normal tissue and patient plasma relative to pooled control plasma from six patients each with Stage I and Stage II/III NSCLC adenocarcinoma. Red, up-regulated; green, down-regulated; grey, high p-values (>0.05) were assigned as missing data. Log2 ratios are presented.
Four of these (miR-25, 32, 142-5p, and 148a) approached significance (p<0.1) and miR-29a was identified at p<0.01 by t-test (Table 3).
Searching miRDB for targets of these seven microRNAs that were up-regulated in Stage II/III compared to Stage I patients resulted in 416 unique targets with a score greater than 80 in the human genome (Supp Table 4). Enrichment analysis of these targets identified five major KEGG pathways: cancer, focal adhesion, ECM-receptor, p53 signalling, and MAPK signaling pathway (Supp Table 5). The highest-scoring cancer-relevant targets are shown in Table 3.
Table 4: microRNAs dysregulated in this study that have been reported to be prognostic or have targets that function in other cancer types. Only in the case of miR-29a and MMP2 (shaded) have the microRNA and target gene been demonstrated to be co-regulated in the other tumor types. *GALNT7 regulation in cervical cancer is associated with miR-214 expression. nd, not determined.
|
microRNA
|
Gene target |
Function |
Tumor type |
Reference
|
|
miR-29a
|
nd |
Invasion |
Gastric |
21 |
|
MMP2 |
Invasion, apoptosis |
Oral squamous |
22
|
|
nd
|
Prognosis |
Colorectal |
23 |
|
nd |
Early detection |
Colorectal |
24
|
|
COL11A1
|
Progression |
Gastric |
28 |
|
COL11A1 |
Cancer-associated stromal cells |
Colorectal |
29
|
|
miR-142-5p
|
MEGF (FAT2) |
Invasion |
Squamous carcinoma |
35 |
|
PSAT |
Proliferation |
Colorectal |
36
|
|
IGF2BP3
|
Prognosis |
Colorectal |
38 |
|
IGF2BP3 |
Prognosis |
Pancreatic ductal |
40
|
|
CHSY3
|
Invasion |
Colorectal |
39
|
|
miR-148a
|
MMP7 |
Invasion |
Gastric |
21 |
|
EIF2C4 |
Metastasis |
Colorectal |
42
|
|
miR-27a
|
GALNT7* |
Invasion, proliferation |
Cervical |
44 |
| miR-140-5p |
SEPT2 |
Proliferation |
Liver |
47
|
DISCUSSION
In this study, we found a consistent down-regulation of microRNAs in tumor compared to adjacent normal tissue and up-regulation of plasma microRNAs from lung cancer patients relative to normal controls. Of the 31 microRNAs included in the TaqMan assay, most were consistently detectable; however five microRNAs had highly variable, often low, values between triplicates, giving a low p-value. Two of these, miR-17-3p and 205, have been reported to have very weak intensities in plasma and serum.18 There was a slightly lower overall expression of tumor microRNAs and corresponding higher overall expression of plasma microRNAs for the youngest patient (L258), but this was not statistically significant. This patient was also the only non-smoker and had the most advanced stage (IIIA). Although biological features of the patient can impact microRNA levels, it would be difficult to draw any valid conclusions concerning microRNA dysregulation based on these features from the results of a single patient.
We were unable to detect any statistically significant changes in tumor tissue microRNAs between Stage I and Stage II/III patients. However, patient plasma did exhibit several microRNAs (miR-25, 27, 29a, 32, 140-5p, 142-5p, and 148a) that could distinguish between patients with Stage I and Stage II/III lung adenocarcinoma, with miR-29a being the most statistically significant (p<0.01). Since plasma is a more easily accessible biopsy material, plasma profiles rather than tissue profiles were
used to investigate alterations in cancer pathways.
There is limited information on the expression of these microRNAs and their target genes in other solid tumor types. There have, however, been some studies describing these microRNAs as prognostic biomarkers and others describing the same genes targeted by the dysregulated microRNAs we have identified. These studies mainly include tumors associated with the digestive system (particularly colorectal) and lend support to our findings (Table 4).
Similar to lung adenocarcinoma, expression of miR-29a is also down-regulated in gastric tumors19 and oral squamous carcinoma,20 resulting in increased expression of Matrix Metalloproteinase2 (MMP2) in the latter case. MMP2 degrades extracellular matrix and promotes tumor invasiveness. In Stage II colorectal cancer, down-regulated tissue miR-29a is associated with high recurrence21 and elevated plasma levels are associated with advanced stage.22 Consistent with our results, the miR-29 family has been reported to be down-regulated in NSCLC tumors relative to normal tissue23,24 and up-regulated in serum of early stage lung adenocarcinoma patients who recurred within 24 months.16 We also found miR-142-5p to be up-regulated in plasma of later-stage patients. This microRNA is derived from the same precursor as miR-142-3p, which is significantly upregulated in serum of early stage lung adenocarcinoma patients who recurred within 24 months16 and in plasma of patients with aggressive NSCLC.25
Of the gene targets of differentially regulated microRNAs listed in Table 3, several do not have a known direct role in NSCLC and are not discussed. These include Brain-specific Angiogenesis Inhibitor (BAI3), SNAP25 (involved in neurotransmitter release) and TET2 and TET3 oncogenes (epigenetic regulators in malignant hematopoiesis).26
The remaining gene targets have roles in ECM-receptor interactions, focal adhesion, cell motility and migration, angiogenesis, protein modification, invasion, proliferation, cell signalling and NK-cell anti-tumor immunity (Figure 3).
Figure 3: Cancer pathways affected by microRNAs differentially regulated in NSCLC progression.
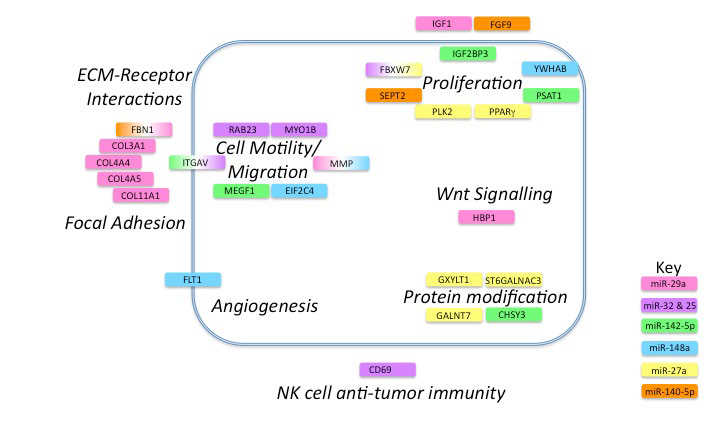
Gene targets are represented by colored boxes with colors indicating microRNA involved. If the gene is targeted by more than one microRNA a gradient of the appropriate color is applied. The tumor cell membrane is represented by a blue line and targets are shown as extracellular, intracellular or membrane-bound. Names of pathways are shown in bold italic font.
This is in agreement with a recent microarray study we performed on the same tumor biopsies27 in which ECM-receptor interactions and components of the focal adhesion pathway were the main elements differentially regulated between early and later-stage lung adenocarcinomas.
miR-29a targets collagens COL3A1, COL11A1, COL4A5, and COL4A4, and Fibrillin1 (FBN1), which participate in integrin signaling, focal adhesion and migration. In gastric cancer, transcripts for several collagens, including COLA11A1, are also up-regulated in malignant compared to pre-malignant tumors28 and COLA11A1 is also up-regulated in cancer-associated fibroblasts of colon adenocarcinomas.29 Insulin-like Growth Factor 1 (IGF1) is a key player in NSCLC tumorigenesis and metastasis,30 initiating cell signaling, promoting mitosis, proliferation and differentiation, and inhibiting apoptosis. HBP1 is a transcriptional repressor that plays a role in Wnt signaling and induces cellular transformation.31 Reduction in tumor tissue miR-29a would result in enhanced IGF-1 and Wnt signalling.
Interestingly, we found two microRNAs (miR-25 and miR-32) that both targeted the same four genes (CD69, FBXW7, RAB23 and MYO1B). As with any biomarker discovery, there is high potential for false positives due to the large number of variables relative to the number of patient samples. Therefore, this overlap in targets suggests that these microRNAs and their target transcripts warrant further investigation as biomarkers in early lung adenocarcinoma. Their higher expression in plasma and concomitant low expression in tumor tissue would promote translation of the hedgehog-signalling factor, RAB23 and the cytoskeletal protein MYO1B, which are necessary for cell motility and metastasis.32 FBXW7 is part of the E3 ubiquitin ligase complex and targets cyclin E for degradation, thus impacting cell proliferation.
microRNAs in the plasma can affect immune cell function. CD69 is a marker of NK-cell activation and a positive regulator of anti-tumor activity.33 It has been shown that administration of several synthetic microRNAs or microRNAs in exosomes from healthy donors to NK-cells and mice bearing tumors resulted in TLR1 activation and increased NF-kB signaling. This resulted in increased NK-cell activation via upregulation of CD69 and increased anti-tumor immunity.34 However, high plasma levels of miR-32 and -25, which repress CD69 expression, would result in reduced NK-cell anti-tumor activity leading to tumor progression.
miR-142-5p targets MEGF1 (FAT2), which mediates migration of human cutaneous squamous carcinoma cells,35 and PSAT1, which has been shown to stimulate proliferation in colorectal cancer.36 miR-142-5p also targets ITGAV and its reduction in tumor tissue would result in an increase in ITGAV. Interestingly, miR-32 and miR-25 also target ITGAV37 and miR-32 is reported to be down-regulated in another lung cancer study.23 CHSY3 and IGF2BP3 are two other relevant targets of miR-142- -5p and both are over-expressed in colorectal cancer patients relative to healthy controls.38,39CHSY3 is responsible for sulfation of glycosaminoglycans on the cell surface and shows elevated expression in colorectal cancer;39 IGF2BP3 has been suggested as a biomarker of poor prognosis in pancreatic ductal adenocarcinoma.40
miR-148a, like miR-29a, is down-regulated in gastric cancer and has a major impact on the expression of MMP7 and invasiveness.19miR-148a also targets FLT1, the receptor for Vascular Endothelial Growth Factor (VEGFR1) and thus could play an important role in tumor angiogenesis. Decreased levels of tumor miR-148a would result in higher levels of FLT1 available for angiogenic signaling. NSCLC tumors expressing FLT1 are more malignant and associated with poorer outcomes.41 Similarly, higher levels of YWHAB, a 14-3-3 protein involved in cell cycle control and inhibition of apoptosis, would result in a more malignant phenotype. EIF2C4 is an argonaute protein in the RNA-induced silencing complex. Argonaute proteins are upregulated in colon cancer and higher levels are associated with distant metastasis.42
Two other microRNAs that showed statistically significant differential expression by SAM (but not t-test) target key genes of importance in cancer progression. miR-27a regulates expression of three enzymes that participate in post-secondary modification of important proteins (GXYLT1, GALNT7, ST6GALNAC3). GXYLT1 glycosylates NOTCH proteins and reduces cancer signalling43 whereas GALNT7 and ST6GALNAC3 glycosylate mucins, thus impacting proliferation, migration, and invasion. Oncogenesis in cervical cancer cells is effected through miR-214 regulation of GALNT7.44 PPAR-γ is a nuclear hormone receptor that is involved in cell proliferation and is over-expressed in many human cancers. It has recently been shown to prevent metastasis in lung cancer cells by inhibiting TGF-β induced epithelial-to-mesenchymal transition.45PLK2 is a serine-threonine protein kinase that is essential for cell division.46 miR-140-5p, like miR-29a, targets FBN1. SEPT2, which plays an important role regulation of cell proliferation through its effect on actin filament formation and has been suggested as a target for liver cancer therapy,47 is also a target of miR-140-5p. The growth factor FGF9 is also involved in cell proliferation and can synergize with the EGFR oncogenic pathway in lung adenocarcinoma48 leading to recurrence.49
The use of microRNAs as clinical biomarkers is increasingly being described in the literature.50 They are highly stable in circulating blood plasma and can be readily isolated. They are tissue-specific and their expression can be dysregulated in response to physiological changes such as disease. Furthermore, they play a key role in differentiation, proliferation and apoptosis making them ideal diagnostic, prognostic and predictive biomarkers in cancer. Although there are some published studies describing microRNA signatures associated with recurrence in lung cancer, they tend to include lung cancer patients with multiple NSCLC tumor types and from multiple stages.15,51,52 It is well known that different types of NSCLC (such as squamous and adenocarcinoma) express different miRNAs. In our smallscale study, we have restricted our comparison to only adenocarcinomas of Stage I and Stage II/III in order to more accurately identify plasma microRNAs likely to be involved in recurrence in early-stage patients. Of the six Stage I and six Stage II/III patients studied, recurrences only occurred in two patients both of whom were Stage II.
CONCLUSIONS
In conclusion, our study indicates that several microRNAs that distinguish between Stage I and Stage II/III patients are involved in the regulation of important pathways in cancer biology including focal adhesion, ECM-receptor interactions, p53 signalling, and MAPK signaling pathway. Because of the small number of samples in this study, their role in lung adenocarcinoma should be verified in a larger patient cohort. Their presence in plasma provides a readily accessible biofluid for clinical testing and the significant up-regulation of miR-29a indicates it could be a promising biomarker.
ACKNOWLEDGEMENT
We gratefully acknowledge the technical assistance of Jeffrey Gallant and Evelyn Teh, and bioinformatic analysis by Susanne Penny, National Research Council (NRC). We thank John Fris, database manager of the Lung Tumor Bank at Capital District Health Authority, for collating clinical annotations. Funding was provided by NRC and Dalhousie University Departments of Pathology and Surgery.








