INTRODUCTION
Mastitis is an infection of the mammary gland caused by pathogenic bacteria that infiltrate the gland. Furthermore, teat canal injury causes physical, chemical, pathological, and bacteriologic alterations in glandular tissues and milk composition. It is one of the most serious and commercially significant illnesses in the dairy industry worldwide.1
Mastitis results in increased economic losses in milk production because it inflames the afflicted quarters. Bovine mastitis decreases milk output, raises the rate of culling, increases medical expenses, and frequently results in severe infections that are fatal.2 Additionally, it can be considered a classic example of the ‘zoonotic spillover’, where a zoonotic pathogen promotes a ‘spillover transmission’ of infection that spreads from the reservoir host to the environment.3
Mastitis can be categorized in a variety of ways. According to the source, mastitis cases can be classified as either infectious or environmental. Contagious mastitis spreads from other diseased areas, whereas environmental mastitis is brought on by bacteria from the environment, sometimes known as environmental pathogens.4 Contagious pathogens such as Staphylococcus aureus and Streptococcus agalactiae, and less common species like Mycoplasma bovis and Corynebacterium, live on the cow’s udder and teat skin, colonizing and growing into the teat canal.5,6
Environmental pathogens survive in the cow’s environment and enter the udder by propulsion through the teat canal (e.g., during milking, by capillary action, insertion of antibiotic tubes, insertion of teat canulae), passive penetration of the teat canal immediately after milking, E. coli, and Strep. Uberis is an important environmental pathogen, whereas Pseudomonas aeruginosa, other coliforms, Bacillus cereus, yeasts and molds, and Pasteurella species are less common.7
Based on the severity of the inflammation, mastitis can be classified into clinical or subclinical forms. Clinical mastitis is characterized by the presence of obvious signs of infection, including aberrant milk (color changes, the presence of clots, flakes), abnormal mammary glands (tissue color changes, swelling), and changes in the animal’s condition, body temperature, appetite, and hydration level.8On the other hand, subclinical mastitis is defined by the lack of observable clinical symptoms. However, milk quality and production yields are typically negatively impacted.9
Bovine mastitis remains a major prevalent disease in cattle and places a significant economic burden on developing countries due to the lack of problem identification and appropriate intervention measures.10 As a result, accurate disease diagnosis is a critical step between disease aetiology and cure. A cost-effective, dependable, and quick diagnostic instrument is essential for udder health management.11
Diagnostic methods in most clinical laboratories are based on the microbiological culture of milk and biochemical assays for subsequent identification. On the other hand, microbiological culture has several disadvantages. Milk culture may not yield bacteria from subclinically infected glands due to the low quantity of pathogens present when samples are taken. Microorganisms from mastitic milk may not be recognized due to the presence of leftover treatment antibiotics in the examined milk. These methods are both time-consuming and labor-intensive.12
Although methods such as electrical conductivity (EC), California mastitis test (CMT), and somatic cell count (SCC) are available, they can only detect a limited number of pathogens or provide limited information about the infection. These methods, too, are only concerned with determining whether or not mastitis occurred, and their sensitivity and specificity are affected by a multitude of factors.13
Due to the limitations of culture and other conventional methods, molecular techniques like polymerase chain reaction (PCR) have been developed to detect different mastitis pathogens. The development of PCR-based technology provides a promising option for speedily identifying microorganisms. This method detects bacterial infections in hours rather than the days required by microbial culture techniques. PCR can also boost detection levels due to its high sensitivity. Furthermore, PCR may detect bacteria in the presence of preservatives or residual therapeutic antibodies in milk, avoiding the false-negative result caused by a lack of bacterial growth, which is a significant disadvantage of the cultural method.14,15
In recent years, novel emerging diagnostic technologies, including infrared thermography, biosensors, and nanotechnology methods, as well as microfabrication of portable, usually digital devices possessing superior diagnostic features, have been scoped for improving the diagnosis of mastitis both at microbial and biomarker levels.9,16
The early, quick, and accurate identification of mastitis is made possible by advancements in mastitis diagnostics. Technology development has caused a significant shift from the use of conventional diagnosis, which has lower specificity and/or sensitivity, to highly measurable, quick, and reliable molecular diagnosis, which has a high degree of accuracy. Therefore, the objective of these papers is to review current diagnostic methods for bovine mastitis.
DIAGNOSTIC METHODS
The identification of etiological agents is necessary for controlling the disease, reducing the risk of chronic infections, and targeting antimicrobial therapy. The suitability of a detection method for routine diagnosis depends on several factors, including specificity, sensitivity, cost, time in producing results, and suitability for largescale sampling of milk.16
Diagnostic tests are frequently evaluated using the scientific standards of specificity and sensitivity. The majority of published test assessments are laboratory-based rather than farmbased tests that should be validated in the host species and under the conditions where they are intended to be utilized.17 Comparing the test results to a reference test or “gold standard”, estimates of scientific criteria are frequently obtained. Latent class analysis, or no-gold standard comparison, is also utilized because no test is flawless.18
Monitoring udder health performance is impossible without reliable and affordable diagnostic methods. Thus, there is a constant need to improve these methods, be it in terms of accuracy, cost, or convenience. Thus, early detection of subclinical mastitis cases is diagnostically important to allow for quick intervention to prevent further development of the condition in individual animals and outbreaks among herds.16
Basic procedures for diagnosing clinical mastitis include udder and teat palpation and visualization of blood, clots, or flakes in the milk. Subclinical mastitis detection requires more advanced approaches, such as somatic cell evaluation, plate-culture procedures, power of hydrogen (pH), electrical conductivity, enzyme activity, molecular diagnostic tools, and biosensors.19,20
COMPARISON OF DIAGNOSTIC TECHNIQUES
Clinical Diagnosis Methods
A physical examination of the mammary gland is important for the successful detection of mastitis. It requires first information about the animal’s general condition, which is the result of a feeling of the animal, feeding, possible trouble or diseases, but also the age and milk productivity of the individual cow. It is important to view the shape, size, consistency, and contour of the udder properly. A detailed examination of the teat and teat orifices should be made to assess inflammation, hotness, pain, swelling, and loss of function.21
Additionally, the firmness of the udder can also serve as an indicator of bovine mastitis. The firmness of the infected and normal udders before and after milking is checked, and a considerable increase in the firmness of the infected udders after milking can be observed. Udder firmness and its relationship with the disease led to its application as a predictive indicator of mastitis.22
Direct Microscopy Examination Method
Microscopically, an examination can be used in the diagnosis of bovine mastitis. The milk sample can be centrifuged, and the deposit can be dyed. A Gram stain is routinely used to detect Grampositive pathogens such as Staphylococcus and Streptococcus and will also reveal yeasts such as Candida albicans that are stained deeply by crystal violet. A modified Ziehl Neelsen-stained smear can be made if Nocardia asteroides is suspected, and a Ziehl Neelsenstained smear is used in rare cases when acid-fast bacteria, such as Mycobacterium fortuitum or Mycoplasma bovis, are present.23
Direct microscopic examination is the reference method that can be used to discriminate between gram-positive and gramnegative bacteria and enables examinations of bacterial morphology, which can provide valuable information. However, it has many limitations. It is time-consuming, skilled labor is required, and it is difficult to differentiate between cells and cytoplasmic particles.24
Conventional Methods
Strip cup test:
It is one of the side tests and can be used for determining the presence of clinical mastitis through the detection of visible particles of milk. It is a simple and effective approach to detecting cows with clinical mastitis. Any layman can make use of this strip cup. In this test, an enamel plate divided into four strip cups is used, and the bottom of the plate is black in color so that the milk flakes are easily observed by tilting the cups at an angle.5 Abnormal milk is usually discolored, watery, or contains flakes and shreds. Similarly, udder tissue can be examined for visible abnormalities, namely swelling, redness, and pain.25
California mastitis test:
The CMT is a simple, rapid screening test based on the estimation of the number of somatic cells in the milk sample.26The somatic cell population is primarily made up of leucocytes (75%), whereas epithelial cells make up 25%.27 The CMT qualitatively estimates the number of somatic cells in milk secretions and is performed by mixing 2 mL of milk sample with 2 mL of the CMT detergent, which dissolves cell walls and releases deoxyribonucleic acid (DNA). The more cells in milk, the more DNA is released, and the thicker the mixture, indicating the presence of high SCC. Hence, SCC can be scored based on the degree of thickening or gel formation.28 According to European Union countries, bulk milk SCC indicates the presence of subclinical mastitis (SCM) in the herd when it counts above 400,000 cells/mL, >200,000 cells/mL SCC for individual cows of composite milk, and individual quarter SCC >50,000 cells/mL (Figure 1).29
Figure 1. On the Left, the more Gel the more Mastitis has been Found within the Milk Ona Zumba30
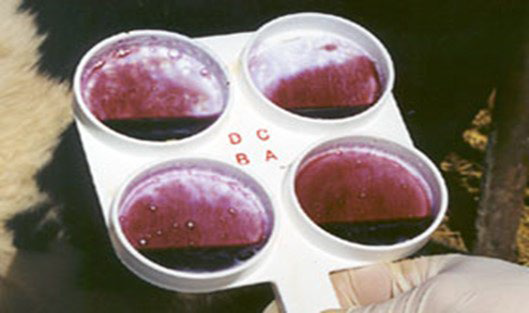
The use of CMT is not recommended for the detection of mastitis four days after calving. However, the test is of high value for monitoring the success of therapy based on the estimation of SCC following treatment. CMT has been less accurate (87.4- 90.8%), less sensitive, and more specific than other tests like SCC, which have shown a sensitivity of 94.9-99.5% and a specificity of 48.1-87.1%.31 Besides, CMT is a time-consuming process, thus not suitable for large numbers of samples, especially in large farms.32
Wisconsin mastitis test:
The Wisconsin mastitis test (WMT) is essentially a laboratory examination that is typically done on samples of bulk tank milk. The same kind of reagent is utilized in WMT and CMT. While the test result reaction in WMT is measured, it is qualitatively evaluated in CMT. A measured amount of reagent and milk are mixed to perform the test. Then, for 8 to 10 seconds, the milk and reagent are combined. The mixture is emptied for 18 seconds before being placed back upright. The fluid level in the tube is monitored after one minute has passed. Typically, WMT scores are measured in millimeters and used to estimate the typical number of somatic cells present. There is a relationship between WMT scores and SCC.33
Somatic cell count test:
SCC is one of the conventional methods used to detect the presence of mastitis in the herds and to assess the sanitary quality of milk. A high somatic cell count number in raw milk indicates not only that the cows have mastitis but also information on metabolic changes in the milk, all the way up to production losses.34
A cow is considered healthy if the SCC of its milk is less than 500×103 cells/mL. Otherwise, the cow is infected with mastitis. Mastitis degrees are classified into four grades: negative, weakly positive, positive, and strongly positive if the SCC is less than 500×103 cells/mL, between 500×103 cells/mL and 1500×103 cells/mL, between 1500×103 cells/mL and 5000×103 cells/mL, and greater than 5000×103 cells/mL, respectively.35
The most accurate relationship between subclinical mastitis and SCC exists at the quarter level, with sensitivities between 30 and 89% and specificities between 60 and 90%.36 Diagnostic sensitivity and specificity for the described method are often compromised by false positives. The SCC parameter can be influenced by many factors, including animal stress, nutrition, stage of lactation, parity, and the quality of the milk sampled.37
Electrical conductivity test:
Mastitis alters the actual ionic dynamics of vascular components due to increased cellular death and a weaker milk-blood barrier. The loss of intracellular potassium causes an increase in the quantity of sodium, potassium, calcium, magnesium, and chloride ions in the blood, while the concentration of potassium ions decreases. These mechanisms change the EC of milk and raise its pH. These differences serve as a diagnostic indicator for differentiating milk with unique properties. The EC of milk has been extensively used for the identification of clinical mastitis due to its simplicity and quickness, with a cost per sample virtually comparable to the cost of the equipment.38
Advanced Molecular Methods
Based on the enzyme test:
The immune response of an animal to an infection and changes in vascular permeability also cause enzymes to be released into milk. While the enzymes involved in inflammation tend to rise, those involved in milk synthesis tend to decline. The phagocyte-produced enzymes multiply rapidly. Nacetyl-B-D-glucosaminidase (NAGase), glucuronidase, and catalase are a few examples of these enzymes.39 Other blood-born enzymes become more active, such as plasminogen, which is locally activated to become plasmin, a proteolytic enzyme that breaks down casein and fibrin40 Typically, enzyme-based diagnostic tests are not accurate since they can differ in other disorders as well as in mastitis-affected cows.41
Power of hydrogen test:
The pH in average milk is between 6.5 and 6.7; however, when a cow has mastitis, the pH rises owing to the alkalinity of the milk. The permeability of blood capillaries improves when the mammary gland becomes swollen, allowing alkaline blood components such as sodium and bicarbonate ions to enter milk, raising the pH of milk to 7.0 in severe clinical mastitis. Beneficial areas with a somatic cell count of <100,000 cells/mL at 37 °C had lower pH values of 6.40 and 6.60, respectively, as well as lower pH values in initial lactation.42
The accuracy and efficacy of pH for the diagnosis of different types of mastitis can be checked by comparing the results of pH testing with other parameters like somatic cell count, electrical conductivity, and CMT scores of the same infected cows. A study was carried out to test the efficacy of these parameters along with the relative oxidative stress in cows affected by mastitis. The study done by Mahapatra in 2018 concluded that the results of other parameters like SCC and CMT scores should also be taken into consideration along with pH values for accurate detection of mastitis in cows.43
Microbial Culture Method
Bacterial culture (BC), carried out according to the internationally agreed standards is considered the reference method in mastitis diagnostics.17 BC is based on culturing a milk aliquot from an aseptically taken quarter milk sample on an agar plate and incubating the plate for 24 to 48-hours. Viable microbial cells can be detected as colony-forming units. Representatives of morphologically different colonies, indicating different species are identified based on standard microbiological schemes.23,44,45
Additional biochemical tests may also be performed if needed. On a range of accessible culture media, the majority of pathogens easily thrive. The growth of particular microorganisms can be aided by the use of appropriate culture media. It is possible to identify pathogens either using milk from a bulk tank, at the cow and quarter level.46,47
Initially, the cultural practices of using conventional media (e.g., nutrient agar or broth) for common bacteria such as Streptococci and Staphylococci and slightly specific media (e.g., MacConkey agar for Gram-negative bacteria) evolved. This progressed to highly specific media, like Mannitol salt agar for Staphylococcus species, Eosin Methylene Blue (EMB) agar for E. coli, and pleuropneumonialike organism (PPLO) medium for Mycoplasma species.32,48
Figure 1 shows that Staphylococcus aureus on nutritional agar displays mucoid colonies in (a); S. aureus growth on mannitol salt agar displays yellow colonies (4,5,6), whereas epidermidis displays pink colonies (1,2,3) (c) S. aureus on blood agar displaying hemolysis; (d) the growth of E. coli on MacConkey agar (lactose fermenting, pink colonies); (e) the growth of E. coli on EMB agar (typical green metallic sheen) sample nos. 2, 4, 5, 6; (f) E. coli showing typical green metallic sheen on EMB agar; (g) Pseudomonas on nutrient; (h) Pseudomonas on MacConkey agar showing mucoid colony; (i) Klebsiella on EMB agar showing mucoid colony.49
Rapid culture plates were developed to differentiate between common mastitis pathogens without the requirement for additional enzymatic testing. This enables incubation and identification of bacteria to be undertaken on-farm or within a veterinary clinic. Identification is usually possible within 24-hours. Rapid-culture plates can be used to make decisions about specific treatments. For instance, a cow with little growth might not get an antibiotic. Rapid culture plates come in bi, tri, and quad configurations.50
This means that each of their 2, 3, or 4 sectors contains a unique cultural medium. When used to categorize infections into broad diagnostic categories, like no growth, gram-positive, or gramnegative growth, rapid culture plates are most accurate. However, some products can recognize some infections down to the species level and even the genus level.50
The separation of fungus and Prototheca from mastitic milk is accomplished using a variety of mycological media, including Sabouraud dextrose agar and Pal sunflower seed medium.51
Microbiological milk culturing and biochemical assays are the identification techniques employed in the majority of clinical laboratories. Microbiological culture has several disadvantages, including the potential for bacteria not to be separated from subclinically infected glands due to a low number of pathogens and the existence of leftover treatment antibiotics in the milk obtained, which may impede bacterial development. Consequently, non-culture-based diagnostic strategies are necessary.52,53
Molecular Method
Molecular diagnostic tools have become the gold standard of mastitis diagnosis in the last few years. They enable rapid, qualitative, quantitative, and large-scale diagnosis. In addition to their role in diagnosis, they can identify pathogens at the subspecies level, which is necessary for epidemiological studies. They are increasingly used in mastitis control programs through the identification of suitable candidates for vaccine production and the selection of mastitis-resistant cattle breeds. The present molecular techniques are continuously improved, and new techniques are developed to provide higher sensitivity and specificity and to minimize costs.54
The basic principle of molecular biological techniques is the extraction of template DNA from materials while carefully removing any potential reaction inhibitors, such as somatic cells. To increase DNA concentration and purity while decreasing the cost of purification, new methods are being developed. These methods range from sterilizing culture by boiling it to employing commercial DNA extraction kits, lysis buffers, magnetic beads, or silica columns. Pre-PCR enzymatic treatment of the bacteria considerably improves the PCR’s capacity to detect the suspected pathogen if it is a grampositive organism.55
These techniques can identify the pathogen with greater sensitivity and specificity. The rapidity and sensitivity of diagnosis have increased with the development of PCR technology and its many expansions, including multiplex PCR, real-time PCR, and loop-mediated isothermal amplification (LAMP).56 These DNAbased diagnostic techniques have made it significantly easier to manage dairy farms. The genomic sequences of numerous diseases are used in nucleic acid-based detection.9
Polymerase chain reaction method:
PCR is a molecular technique with the widest variety and application in veterinary diagnostics. The strength of this technique is due mainly to its simplicity, sensitivity, flexibility, and ability to produce millions of copies of a target DNA Salisu et al.57 PCR is an in vitro amplification method that uses heat-stable polymerase, oligonucleotide primers that are specific to the target DNA sequence, and unique organism-specific target DNA sequences. The chosen primers ought to have an exclusive sequence that binds specifically and selectively to the DNA target sequence that has already been identified. The primers may be made to identify the organisms at the subspecies level or to distinguish between individuals of the same species. In doing so, the primers enable the measurement and amplification of specific sequences. The primer target sequence for diagnosing current pathogens must be substantially conserved across all strains of the suspected species to prevent false negative results, but it must be varied.15
Different PCR approaches have been developed to give a quick, accurate, and economical diagnosis of mastitis-causing pathogens. Since the start of the twenty-first century, new technologies, such as conventional, real-time, and multiplex real-time PCR, have been developed for the diagnosis of bovine mastitis.58,59
Milk samples in quarters, pools, and bulk can all be tested using PCR. As infectious milk is diluted (pooled or mixed) with healthy milk, the detection limit’s sensitivity declines. In comparison to pooled or bulk milk samples, quarter milk samples clearly have a greater level of sensitivity and specificity and therefore provide the most reliable data regarding the dominant pathogen in the farm.54 The use of contemporary molecular methods in the investigation of pooled or bulk milk samples can produce accurate results comparable to those obtained when using quarter samples. It can detect one moderate-to-heavy-infected cow with Strep. agalactiae and M. bovis even if the milk was pooled with milk samples from 1000 healthy cows in the herd.60
PCR is more sensitive, rapid, and reliable as compared to the bacterial culturing method for the detection of various mastitis pathogens Cantekin et al61. Certain mastitis pathogens do not grow in bacterial culture or are difficult to detect due to their slow growth rate. PCR assays are quite popular in dealing with such issues by rapidly detecting the pathogen with high specificity. One such study was conducted by Boonyayatra in 2012 to detect three species of Mycoplasma by real-time PCR assay.62
Despite being a promising diagnostic method for mastitis detection and management, the PCR assay has numerous drawbacks, including the lack of specific guidelines or cut-off points for the definition of sample contamination, unlike BC; and its use in developing countries is limited in comparison to developed countries (economic reasons), the potential for false-positive results due to milk carryover (defined as the transfer of a small amount of milk from one cow sample to the next at the time of collection due to the presence of residual milk in the milking unit, milk meter, or milk sampler) the applicability of pre-sampling procedures, and the inability to distinguish between viable and non-viable bacterial cells.11
If samples are transported without preservatives or are left uncooled, even low amounts of some pathogens, such as E. coli, could quickly outgrow the other pathogens, resulting in a falsely high amount of E. coli DNA detected in PCR. A carryover could also be a cause of contamination. One such study was conducted by Mahmmod in 2017 on DNA carryover in cows being examined with PCR for S. aureus. Truly, IMI-negative cows represent the biggest risk for a false positive S. aureus diagnosis due to the carryover from an S. aureus-positive cow milked just before her.63 It is difficult to implement PCR on farms due to sterility standards, the need for advanced equipment, and the demand for qualified personnel. Furthermore, to achieve accurate results, it is necessary to use particular DNA extraction techniques because milk contains known PCR inhibitors, including calcium, fat, and protein. LAMP, a potential alternative to conventional PCR and quantitative PCR (qPCR) techniques, has been suggested as a viable molecular tool for fast on-farm diagnostics.64
Multiplex PCR assay:
Mastitis milk samples can be positively checked, even if no bacterial growth is observed under conventional culture conditions. This may be attributed to the presence of a very low number of pathogens or the inhibitory effect of residual therapeutic antibiotic concentrations on microbial growth.65 On the other hand, multiplex PCR tests are of interest because several pathogens can be tested at the same time. The main drawback with multiplex PCR is that there is competition between different sets of primers for PCR substances like deoxynucleotide triphosphates (dNTPs) and Taq polymerase, which reduces the sensitivity.66
Real-time polymerase chain reaction:
Real-time PCR, also known as quantitative PCR, is a modification of the PCR approach that enables real-time monitoring of the PCR progress Artika et al67. Commercial kits are available for the simultaneous detection of major mastitis pathogens such as S. aureus, Strept. agalactiae and Strept. uberis directly from mastitic milk using a multiplex real-time reverse transcriptase-PCR (RT-PCR) assay with an accuracy of 98%.54
Compared to the use of bacterial culture and regular PCR, RT-PCR has extra advantages. It is not only quicker and more accurate, but it’s also safer for the environment and the workers (no ethidium bromide is used), there’s no need for post-reaction handling (no agarose electrophoresis), and the results are better visualized and digitalized, allowing for data sharing with other teams and documentation. When used to identify mastitis pathogens, RTPCR has a 100% sensitivity and specificity rate.68,69
Loop-mediated isothermal amplification-based method:
LAMP is an established isothermal nucleic acid amplification method developed in the year 2000.70 LAMP is a technique that detects the genomes of the pathogens that cause bovine mastitis instead of a bacterial culture by first purifying the nucleic acids from milk and then detecting them on DNA chips.71 The Toshiba Corporation has reported a technique for detecting DNA chips that utilizes an electrochemically active Hoechst 33258 intercalator and a DNA probe mounted on a gold electrode. The DNA chip is designed for mastitis pathogen detection. While several nucleic acid amplification techniques, including the PCR technique, have been established thus far, the LAMP technique is particularly promising because DNA can be amplified more quickly and specifically under isothermal conditions compared to PCR.70,72
The main advantage of this technology is that, like PCR, it is purported to be faster and highly specific for the target sequence. Its key potential advantage over PCR is that, because it amplifies DNA under isothermal conditions, it might be deployed in the field, requiring only a water bath or heat block for the reaction to proceed.73 The LAMP reaction can be conducted in a water bath or heat block with an ideal temperature range of 60-65 °C, and the assay uses four specifically designed primers capable of recognizing six different sections in the target DNA, making the process very precise. Results can be directly visualized by adding a dye like SYBR green (SG); a change in color due to the presence of magnesium pyrophosphate, a LAMP byproduct, indicates the positive samples.74,75
LAMP tests for common mastitis pathogens such as Staphylococcus aureus, Streptococcus agalactiae, or Streptococcus uberis have been developed and validated.76 However, due to the enormous numbers of DNA amplicons generated during the LAMP assay, visualization of LAMP amplicons by gel electrophoresis is timeconsuming and may raise the danger of cross-contamination between samples.75
Emerging Technology Methods
Infrared thermography-based diagnosis:
Infrared thermography (IRT) is a non-invasive diagnostic tool for assessing changes in skin surface temperature that are influenced by the internal conditions of tissues and organs, accompanied by fluctuations in the amount and rate of capillary blood flow.77 Its diagnostic tools can distinguish both clinical and subclinical mastitis. An innovative method that can be used on-site to accurately and quickly diagnose mastitis is infrared thermography. Its foundation is the temperature difference between infected and healthy udders. The degree of udder infection is assessed using heat images created by udder cameras.78 Infrared thermography was investigated to see if it had a strong correlation with somatic cell count. Similarly, because IRT is a mobile-based application, it is very sensitive and farmer-friendly. It can detect even tiny variations in the temperature of the udder surface, making it useful for early-stage mastitis identification.79
The studies conducted using principal component analysis for physiological and environmental thermohydrometric data correlated with the detection of subclinical and clinical mastitis in Capoeiras in the State of Pernambuco. The authors discovered temperature variations between cows with healthy udders (28.79°C) and those with subclinical (32.66 °C) and clinical (37.82 °C) mastitis, demonstrating that affected animals have udder surface temperatures Silva et al80. For the detection of subclinical mastitis in cows, the diagnostic sensitivity and specificity of IRT are 95.6 and 93.6%, respectively, as compared to 88.9 and 98.9% for CMT. However, using IRT, they could not detect local inflammatory changes of the udder, which appeared earlier than the rectal temperature increase.77
Biosensors based:
Recent advances in biosensor technologies have the potential to match or surpass conventional diagnostics concerning sensitivity, selectivity, accuracy, and cost.81 The conventional methods for pathogen detection and identification are either time-consuming (culture and colony counting) or expensive (molecular approaches). Recent developments in micro- and nanotechnologies have spawned a brand-new category of analytical devices called biosensors. Recent developments in micro- and nanotechnologies have spawned a brand-new category of analytical devices called biosensors. More integrated biosensors are built on improved microfabrication processes and innovative nanomaterials with improved sensing capabilities or connected to biomolecules to function as reporters or signal amplification systems.20
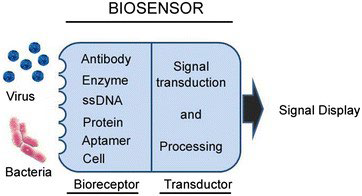
Biosensors for mastitis detection have been developed for on-site testing in an attempt to develop less lengthy approaches to conventional diagnostic methods. Recent advances in nanotechnology and biotechnology have led to the development of analytical tools called biosensors, capable of converting the biological compounds in a sample into electrical signals. These signals can detect the presence of particular cells and markers with high sensitivity upon proper tuning and amplification. The biological element known as the bioreceptor interacts with a physical transducer, called the sensor, and produces a measurable signal that is transformed into data. The most commonly used recognition elements in sensing bacterial contamination include single-stranded oligonucleotides, antibodies, and artificial binding proteins (Figure 3).27,82,83
Figure 3. The Principal of Biosensors Vidic et al84
Biosensors have disadvantages for pathogen detection using milk as a target sample, and in particular for bovine mastitis diagnosis, results are still limited.85 The complexity of the milk matrix is undoubtedly a factor in the restricted availability of fully automated, standalone biosensing technologies for mastitis milk analysis.86 However, further studies are recommended to improve the specificity and sensitivity of this rapid biosensor method.87
CONCLUSION
Mastitis is an inflammation of the mammary gland caused by physical or chemical stimuli, although the majority of the time it is caused by bacteria that invade the udder and generate toxins that are detrimental to the mammary gland. The quicker the disease is discovered, the less harm there will be. Therefore, many efforts are being made to develop reliable diagnostic tools for use in the diagnosis of mastitis, such as conventional methods (strip cup test, California mastitis test, Wisconsin mastitis test, somatic cell count test, and electrical conductivity), molecular methods (polymerase chain reaction), and emerging technology (infrared thermography and biosensors). However, clinical diagnosis, microscopic examination, conventional methods, and microbial culturing are still in use. Microbial culture-based methods are the gold standard for identifying mastitis pathogens and are frequently used. The molecular methods are partially established in advanced laboratory settings and hold the promise of faster and more sensitive results, though they are still expensive to implement in developing countries like Ethiopia. In recent times, emerging technology has helped to detect mastitis with greater sensitivity and rapidity, but it is still limited in veterinary practices.
Based on the preceding conclusion, the following recommendations
are made:
• It should be better to adopt and employ advanced diagnostic methods, such as advanced molecular tests and emerging technology.
• The government should build additional national laboratories and research centers to provide mastitis tests for farmers as a valuable tool for mastitis diagnosis and management.
DATA AVAILABILITY
All the datasets generated or analyzed during this study are included
in this manuscript.
AUTHORS’ CONTRIBUTIONS
GD and TN collected the data, designed the study, interpreted the data, and drafted and wrote the manuscript; IAK and ADA edited and revised the manuscript, and searched the references. All authors have approved the submission of the final manuscript.
FUNDING STATEMENT
The current study was conducted without the support of funding sources.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.







