INTRODUCTION
Haemophilus influenzae (Hi) is a gram-negative coccobacillus bacterial species which contain encapsulated and non-encapsulated strains. There are 6 encapsulated strains (a-f), classified on the basis of their antigenically-distinct capsular polysaccharides. The non-encapsulated strains, termed non-typeable (NTHi), do not possess a capsule and are genetically more diverse than the encapsulated strains.1
Both types of strains cause a variety of diseases in children and adults. Of the encapsulated strains, type b (Hib), is the most virulent and can cause bacterial meningitis and other invasive infections in children under the age of 4-years. The NTHi strains are, with S.pneumoniae, the leading cause of acute otitis media (AOM) in children. Together, they account for around 80% of bacterial otitis media cases.2,3 NTHi has also been implicated in causing exacerbations in chronic obstructive pulmonary disease (COPD) which is currently the third leading cause of death worldwide and expected to be the leading cause of death by 2030.4 As in otitis media (OM), of the bacteria isolated S. pneumoniae, M. catarrhallis and NTHi are the 3 most popular.5
There are currently a number of effective licenced vaccines against Hib.6 These vaccines are comprised of the Hib capsular polysaccharide, polyribosyl-ribitol-phosphate, conjugated to various protein carriers (tetanus toxoid, non-toxic mutant of diphtheria toxin, or meningococcal outer membrane proteins) to overcome the weak immune response to capsular polysaccharide in children younger than 18-months of age. However, for NTHi, several vaccine candidates, mainly outer membrane proteins which are highly conserved between NTHi strains have been considered. These include outer membrane proteins (OMPs) such as P4,7 P6,8 lipooligosaccharides (LOS),9 Adhesins 1 and 210 and protein D.11 All of these OMPs have been shown to be immunogenic in previous studies. However, some OMPs, including Adhesins 1 and 2 (which are high molecular weight proteins that the bacteria use to bind to mucin in the respiratory tract), are able to be down regulated via phase variation to evade immune responses. The lack of constitutive expression of such proteins makes them poor vaccine candidates to be considered effective against all strains of Hi.12 Of the OMPs, protein D is an attractive target for a vaccine because it is highly conserved between strains with limited genetic drift13 and is expressed on all 127 known Hi strains, both encapsulated or not. The hpd gene which encodes protein D13 varies by only a few nucleotides between all known strains of Hi. This low-level of variation is seen even during prolonged infections where the bacterium is under immunological pressure.14 As no protein D mutations have been documented, it is likely that this protein serves an important function within the bacterium. Protein D has been prepared as lipidated and non-lipidated form and preclinical studies showed that protein D antibodies were able to induce 34-38% protection against NTHi otitis media in the chinchilla model.15
A ten-valent conjugate vaccine (Synflorix® from GlaxoSmithKline (GSK), London, UK) has been developed and licenced for use against pneumococcal invasive diseases. The vaccine uses protein D as a carrier for pneumococcal capsular polysaccharides of different serotypes. Eight serotypes are conjugated to protein D and the remaining two are conjugated to either diphtheria toxoid (DT) or tetanus toxoid (TT). Data from clinical trials with Synflorix® showed promising efficacy against NTHi as well as S. pneumoniae. The vaccine efficacy against episodes of otitis media caused by NTHi was reported to be 35%.11
As the NTHi strains lack capsular polysaccharide, immunisation with Hib conjugate vaccine cannot protect against infection with NTHi.16,17 On the other hand, although protein D is expressed in all Hi strains, including Hib, its effectiveness as a vaccine candidate against Hib strains has not been evaluated.
The aim of this study is to investigate the potential of recombinant protein D as a vaccine candidate against both NTHi and Hib. The immunogenicity of recombinant protein D and the protective activity of the induced immune sera will be evaluated in vitro against Hib and NTHIi clinical isolates, using the bactericidal assay and in vivo against Hib Eagan strain, using an infant rat Hib bacteraemia model.
MATERIALS AND METHODS
Expression, Purification and Characterisation of Recombinant Protein D
Expression and purification of protein D:
The genomic DNA of Hib strain CMCC58547 (which is used for the production of Hib vaccine; source) was extracted to amplify the DNA encoding protein D (hpd) by PCR, using a pair of primers: (P1: 5’- AGCACATATGAGCAGCCATTCATCA-3’ and P2: 5’-GAGGAAGCTTATTTTATTCCTTTTA-3’).
Escherichia coli strain M15 and BL21 (DE3) (Novagen, WI, USA) were used for the plasmid construction and protein expression, which were cultured in luria broth (LB) medium at 37 °C. The PCR product was ligated with the pET30a (+) vector (Novagen, WI, USA) at restriction sites Nde I and Hind III. The constructed plasmid was designated pET30a(+)/hpd and transformed into E.coli BL21(DE3). The cloned DNA sequences were verified by sequencing analysis. Expression of the recombinant protein D was induced by the addition of isopropyl-β-D-1-thiogalactopyranoside (IPTG) to E.coli BL21(DE3) carrying the plasmid pET30a(+)/hpd at a final concentration of 0.2 mM. The expressed protein was purified using ion-exchange chromatography and hydrophobic chromatography in tandem with AKTA purifier 100 system (GE Healthcare, Chicago, IL, USA). Briefly, after induction with IPTG, the cells were collected by centrifugation and sonicated on ice bath. The inclusion bodies containing the recombinant protein were separated by centrifugation at 12,000 g for 15 min at 4 °C and solubilized in the binding buffer (6 M urea, 20 mM Tris at pH 9.5). The protein was loaded onto a Q Sepharose Fast Flow column (2.6 cm×20 cm; GE Healthcare, Chicago, IL, USA) and eluted with an increasing concentration of NaCl (0-1 M) in the binding buffer. The main eluate was collected and ammonium sulphate added at 0.5 M for the next step of purification with Phenyl Sepharose 6 Fast Flow column (2.6 cm×20 cm, GE Healthcare, Chicago, IL, USA). The target protein was eluted with the buffer solution (50 mM PB, 6 M urea at pH 7.0) containing a decreasing concentration of ammonium sulphate (0.5-0 M). The recombinant protein D was refolded by gradually decreasing the concentration of urea to 0.5 M by dialysis for 48 h. The purity of the recombinant protein was estimated by SDSPAGE and gel densitometry analysis, and protein concentration was determined by the Lowry method as described previously.18
Protein identification using LC-MS/MS analysis:
Twenty (20) µg of the purified protein D was used for in-solution tryptic digestion according to the method described by Whiting et al.19 Briefly, the sample was denatured, reduced and alkylated prior to overnight digestion with trypsin. The resulted digest was subjected to a U3000 direct nanosystem coupled with a nanoelectrospray and LTQ-Orbitrap Discovery mass spectrometer (Thermo Fisher, MA, USA) as described previously.20 The technical triplicates of the sample were performed. Data analysis was carried out using PEAKS 8 (Bioinformatics Solutions Inc., Ontario, Canada) with precursor and fragment error tolerance as 10 ppm and 0.6 Da, respectively. Deamidation (NQ), carbamidomethylation (C), oxidation (M) and pyro-Glu (Q) were set as variable modifications. Three raw files were searched against a database containing the sequence of recombinant protein D (without signal peptide), and the Swiss-Prot all proteins as background.
Expression of protein D on Hib and NTHi: The relative level of protein D expression on Hib and NTHi isolates was evaluated using quantitative western blotting. In brief, 50 µg of total protein extract quantified by BCA assay (Thermoscientific, MA, USA) was separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes using a semi-dry western transfer apparatus (Thermoscientific, MA, USA). The membrane was initially stained using pierce reversible protein stain (Thermoscientific, MA, USA) to be used for loading control. Nonspecific binding sites on the membranes were blocked by incubation in 5% skimmed milk (Fluka®, Buchs, Switzerland) in TBST for 1 hat RT. After the blocking step, the blot was incubated with antiprotein D mouse serum followed by horseradish peroxidase (HRP)- conjugated goat anti-mouse IgG (Thermoscientific, MA, USA) for 1h at RT. The blot signal was then detected using an enhanced chemiluminescence (ECL) detection kit (Thermo scientific, MA, USA). Relative quantification of protein D expression was achieved through densitometry analysis of protein D bands normalized to total protein stain loading control.21,22
Endotoxin content:
The endotoxin content of the purified protein D was determined using an in-house Limulus Amebocyte Lysate (LAL) assay. Protein D was reconstituted in 0.5 ml of Tris-HCl (0.25 M, pH 9) and mixed with LAL (Lonza) which reacts with bacterial endotoxin or lipopolysaccharide (LPS). The presence of endotoxin results in coagulation of the mixture which can be quantified relative to a reference LPS titrated in the same assay.
Haemophilus influenzae strains:
Four clinical isolates of NTHi, 3 from invasive disease cases (Haemophilus Reference Unit, Public Health England; PHE, UK) and one from OM cases (ATCC, LGC group Teddington, UK), were used in the study. In addition, three clinical isolates of Hib were also used in this study (Haemophilus Reference unit, PHE, UK).
Animals Immunisation
Female BALB/c mice (6-8-weeks: Charles River; 5-8 per group, as indicated for each experiment) were immunised subcutaneously on days 0 and 28 with various doses of protein D with or without adjuvant. Animals were bled on day 42 and sera collected and stored at -20 °C until used.
In vivo Protection Against Haemophilus influenzae Type b Challenge
Female and males (1:1 ratio) Sprague Dawley (Charles River, MA, USA) infant rats (3-5-days old; 10 per group) were injected intraperitoneally (IP) with 100 μl of immune serum diluted 1:1 or 1:25 with PBS or with 100 μl of normal rat serum as a control. After 24 h rats were challenged IP with 104 CFU of a clinical isolate of Hib (Eagan strain; Hib 1) grown to log phase in Muller Hinton broth. Blood was collected by cardiac puncture 24 h later and cultured on chocolate agar plates for 24 h to assess the presence of bacteraemia.
All animal studies were conducted according to the Animals (Scientific Procedures) Act 1986, under UK Home Office project licence PPL80/2634.
ELISA for Quantification of Anti-Protein D IgG and IgG Subclasses
Ninety six (96) well Nunc Maxisorbmicrotitre plates (Gibco, MD, USA) were coated with 100 µl/well of 2.5 μg/ml Protein D in carbonate buffer (pH 9.6) for 2 h at 37 °C and then incubated overnight at 4 °C. Next day, the plates were washed with PBS/0.05% Tween20 (Sigma, MO, USA) and blocked with 100 µl/well of assay diluent [AD; PBS/1% BSA (Sigma, MO, USA), 0.3% Tween20, 0.01 M EDTA (Fisher Scientific, MA, USA)] for 1 h at 37 ºC. AD was then discarded and 11 serial two-fold dilutions of the immune sera in AD were prepared in the plate, starting at 1/50 dilution (in 100 µl volumes) and plates were incubated for 90 min at RT. Binding of protein D antibodies was detected with 100 µl/well of 1/500 dilution of biotin-conjugated goat anti-mouse IgG (Sigma, MA, USA) and plates incubated at RT for 90 min. This was followed by 1 h incubation at RT with 100 μl/well of streptavidin-horseradish peroxidase diluted 1/2000 (Sigma, MA, USA). One hundred (100) µl/well of substrate solution (Ortho-phenylenediamine/H2 O2 ; Sigma, MA, USA) was added and plates incubated at RT in the dark for up to 30 min. The enzymatic reaction was stopped by adding 50 µl/well of 3 M HCl and the optical density was measured at 492 nm using a Multiskan MS plate reader (Labsystems, Helsinki, Finland; Thermo Life Sciences, MA, USA). Each serum sample was tested in duplicates on separate plates and data presented as mean endpoint titre for IgG (reciprocal of serum dilution giving OD>mean blank+2 SD). To detect the level of IgG1 and IgG2a anti-protein D antibodies, ELISA was performed as above, except for the use of isotype/subclass specific detection antibodies: goat anti-mouse IgG2a-HRP (Invitrogen, CA, USA), or goat antimouse IgG1-HRP (Southern Biotech, AL, USA). Data for IgG1 and IgG2a anti-protein D are presented as relative concentration (µg/ml), calculated from standard curves generated from serially diluted, commercial mouse IgGI and IgG2a myeloma proteins of known concentration (Sigma, MO, USA), tested on plates coated with anti-IgG1 or anti-IgG2a instead of protein D, using an inhouse parallel line bioassay programme (CombiStatsTM, Strasbourg, France) which relates the logarithm of the assay responses (OD) to the logarithm of the sera dilution, using at least 3 points on the linear portion of the titration curve.
Bactericidal Assay
Heat inactivated (55 °C for 30 min) anti-protein D immune sera were serially diluted in 40 µl volumes in the first 10 columns of a flat-bottom 96-well microtitre plate (Falcon, Mexico, USA), in assay buffer [Hanks balanced salt solution and 10% Foetal bovine serum (Gibco, MD, USA)]. Assay buffer was added to the last 2 columns which serve as negative control for the assay.
The bacteria to be tested were grown to log phase in Mueller-Hinton broth and then diluted in assay buffer to 1×104 CFU/ml before use in the assay. Twenty (20) µl of bacterial culture were added to each well, followed by 30 µl of 1:1 diluted baby rabbit complement (Mast Group, Ohio, USA), which was added to all wells, except the last column which serves as a negative control. The plate was incubated at 37 °C for 1 h in a shaking incubator (100 rpm). At the end of the incubation, 3×10 µl from each well were spotted onto pre-warmed chocolate agar plates which were tilted gently to allow the bacteria suspension to run along straight lines. The agar plates were incubated overnight at 37 °C and the number of colonies counted the next day. Each serum dilution was tested in triplicate wells and the results were presented as a bactericidal titre, which is the serum dilution that caused 50% killing of the bacteria.
STATISTICAL ANALYSIS
Comparison between the groups was done by the Mann Whitney two sample rank sum test for the immunogenicity studies and by the student’s t-test for the bactericidal assay. A Bonferroni correction was used to adjust the significance level for multiple comparisons with a control group.
RESULTS
Expression and Characterization of Recombinant Protein D
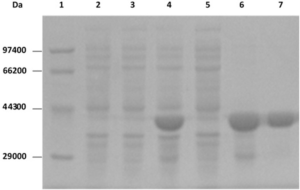
To generate recombinant protein D in E.coli, the respective gene was amplified from a Hib vaccine strain and cloned into a protein expression vector. The expression plasmid was constructed, and the DNA sequence confirmed by DNA sequencing. Following Isopropyl β- d-1-thiogalactopyranoside (IPTG) induction, the recombinant protein (346aa) without 18aa signal peptide and without affinity tag was successfully expressed in E.coli as inclusion bodies. The induced protein band corresponded to the predicted size of 40-kDa on SDS-PAGE (Figure 1), which accounted for 57% of the total cellular proteins analysed by using Image Master VDS Software (GE Healthcare, IL, USA). The recombinant protein D was purified using ion-exchange chromatography and hydrophobic affinity chromatography methods, yielding a highlevel of purity (>95%; Figure 1).
Figure 1. SDS-PAGE of Recombinant Protein D Isolated From E. coli Expression System
Identification of protein D using LC-MS/MS analysis indicated the presence of 35 unique peptides and an amino acid sequence coverage of 62% (Figure 2).
Figure 2. Identity of Protein D Specific Peptides by Mass Spectrophotometry

Relative Expression of Protein D on Haemophilus influenzae Strains
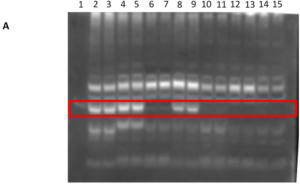
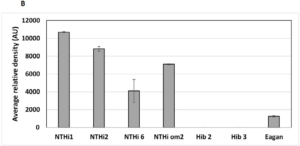
A quantitative western blot analysis was performed to enable a relative comparison of the level protein D expression on the different Hi strains used in this study. Upon analysis of the whole protein-normalised western blot signal, higher protein D expression levels were observed in the NTHi strains compared to Hib strains (Figure 3A, lanes 2-9). Protein D expressed by Hib strains 2.2 and 2.3 were below the detection limits of the assay performed and the Eagan strain expressed relatively much lower protein D levels (Figure 3A, lanes 10-15) compared to all 4 NTHi strains.
Figure 3. Expression of Protein D in Various Haemophilus influenzae Strains
The endotoxin content of the recombinant protein, determined by the LAL assay, was very low (10 IU/mg of protein), suggesting its suitability for use in animal immunisation studies.
Expression of Protein D on the Different Haemophilus influenzae Strains
A quantitative western blot analysis was performed to enable a relative comparison of protein D levels among the Hi strains used in this study. Upon analysis of the whole protein normalised western blot signal, higher protein D expression levels were observed in the NTHi strains compared to Hib strains (Figure 3B). Protein D expressed by Hib strains 2 and 3 were below the detection limits of the assay performed and the Eagan strain expressed relatively lower protein D levels.
Immunogenicity of Protein D in a Mouse Model
Immunogenicity of various doses of protein D and kinetics of the immune response:
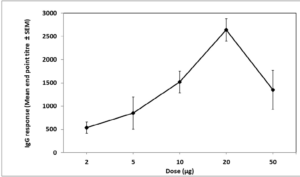
Immunisation of mice with 2 µg of protein D on days 0 and 28 induced a weak IgG antibody response (Mean titre=538) which was further increased with increasing the dose to 5, 10 and 20 µg in a dose-dependent manner, reaching a maximum titre of 2,640 (Figure 4). Further increase in the immunisation dose to 50 µg yielded a slightly lower response (mean titre=1,348). Although the difference between 2 consecutive doses was not always significant due to the high variability in the response between individual animals, the response was linear in the range between 2 and 20 µg doses (R=0.968).
Figure 4. Immunogenicity of Protein D
Effect of Various Adjuvants on Protein D Immunogenicity
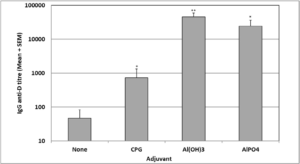
Investigation of the effect of various adjuvants on the anti-D antibody response showed that all three adjuvant formulations: Al(OH)3, CPG and AlPO4 had significantly increased the antiprotein D IgG level when compared to the IgG level following immunisation with protein D on its own (Figure 5). However, the level of adjuvanticity varied for the different adjuvants, with CPG being the weakest adjuvant, inducing a 46 fold increase in the antibody response (p<0.05) followed by AlPO4 (524 fold; p<0.05) and Al(OH)3 (969 fold; p<0.01).
Figure 5. Effect of Various Adjuvants on Protein D Immunogenicity
IgG Subclass Distribution
IgG subclass distribution in sera induced by immunising mice with protein D in the presence or absence of adjuvant was evaluated for IgG1 and IgG2a. Data displayed in Table 1 shows that IgG1 was more dominant than IgG2a, suggesting that the immune response is more skewed towards a Th2-type response. The Al(OH)3 group had the highest levels of IgG1 (308.9 µg/ml) compared to IgG2a (0.038 µg/ml) with an >8,000 fold difference between the two subclasses. This was followed by the AlPO4 group which had a 7,000 folds difference between the IgG1 (146.3 µg/ml) and IgG2a subclasses (0.021 µg/ml). The CPG and the no adjuvant groups had similar IgG1:IgG2a ratio (16) but with CPG group inducing a much higher response (0.48 and 0.029 µg/ml for IgG1 and IgG2a, respectively).
| Table 1. IgG1 and IgG2a Concentration in Sera of Mice Immunised with Protein D and Different Adjuvants |
|
Adjuvant
|
Relative Concentration (µg/ml) |
IgG1: IgG2a ratio
|
|
IgG1
|
IgG2a
|
|
No adjuvant
|
0.068 |
0.004 |
16 |
|
CPG
|
0.48 |
0.029 |
16 |
| Al(OH)3 |
308.85 |
0.038 |
8,127
|
| ALPO4 |
146.28 |
0.021 |
6,966
|
| Sera samples generated after two immunisations with protein D in the presence or absence of adjuvant were analysed for anti-protein D IgG2a and IgG1 concentrations relative to a commercial mouse IgG1 and IgG2a myeloma protein reference standards. |
Functional Activity of Protein D Antisera Against NTHi and Hib Clinical Isolates
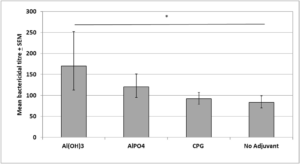
To evaluate the functional activity of antibodies generated by recombinant protein D against NTHi, pooled sera samples from immunised animals were tested in serum bactericidal assays against a single NTHi clinical isolate (NTHi6, obtained from an invasive disease case). Immune sera of animals immunised with protein D, in the presence or absence of adjuvant, were bactericidal against the NTHi isolate used, with mean bactericidal titres ranging from 83-170 (Figure 6). Sera from mice immunised with Al(OH)3 – adjuvanted protein D were significantly more bactericidal than sera from mice immunised with protein D alone (mean bactericidal titr e=170 vs 83; p<0.05). AlPO4 adjuvant slightly increased the bactericidal titre but this increase was not statistically significant (120 vs 83; p>0.05) and mice immunised with CPG-protein D or with protein D alone had lower and very similar serum bactericidal titres of 92 and 83, respectively. A pool of non-immune sera was used as a negative control in the assay and showed no, or very low, non-specific killing, indicating that the bacteria killing was mediated by protein D antibodies. To ensure that the bactericidal activity of protein D antisera covers a range of NTHi and also Hib strains, 3 more clinical isolates of NTHi obtained from invasive and non-invasive disease cases and 3 clinical Hib isolates from invasive disease cases were also evaluated for susceptibility in the bactericidal assay, using immune serum from mice immunised with Al(OH)3 –protein D (Table 2). Protein D antisera had bactericidal activity against all the NTHi isolates tested with no significant difference in the bactericidal titre against the different isolates (p>0.05). Mean bactericidal titres ranged from 37 for isolate OM2, to a maximum of 79.5 for isolate 2. No relationship was found between the original site of isolation of the bacteria and the bactericidal titre.
Figure 6. Bactericidal Titres of Sera Samples, Induced by Immunisation with Protein D in the Presence or Absence of Different Adjuvants
| Table 2. Bactericidal Titres of Protein D Antisera Against Different NTHi and Hib Clinical Isolates |
|
Isolate
|
Number of Assays |
Mean Bactericidal Titre+SEM |
Source |
Isolated from
|
| NTHi 1 |
3
|
36.3+13.1
|
PHE |
Invasive case (not-specified) |
| NTHi 2 |
6 |
79.5+18.5
|
PHE |
Invasive case (not-specified) |
| NTHi 6 |
11
|
47.4+10.3
|
PHE |
Invasive case (not-specified) |
| NTHi OM2 |
5
|
37.3+9.1
|
LGC |
Otitis media |
| Hib1 |
5
|
24.6+2.3
|
PHE |
Bacteraemia/Meningitis |
| Hib2 |
4 |
28.0+12.1 |
PHE |
Epiglottitis |
| Hib Eagan |
3 |
20.0+5.0 |
PHE |
Meningitis |
| Comparison of the bactericidal titres of protein D antisera against a range of NTHi and Hib isolates. Each assay for each isolate was repeated at least 3 times. Data is presented as mean bactericidal titres +SEM. Data analysis was done by Student’s t-test to compare between the serum bactericidal titres against the various NTHi or Hib isolates. |
When Hib isolates were used as the targets in the killing assay, protein D antisera were found to be able to kill all Hib isolates to similar extents with bactericidal titres ranging from 20-29. The difference in bactericidal activity between the Hib isolates was not significant and was smaller than between the NTHi isolates. However, the mean bactericidal titres against Hib isolates were lower than those against NTHi isolates. Similar to NTHi isolates, there was no relationship between the original site of isolation of the Hib isolates and the bactericidal titre.
In vivo Functional Activity of Protein D Antisera Against Hib Bacteraemia
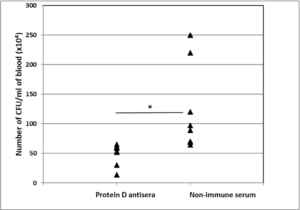
To test if protein D antibodies can kill Hib bacteria in vivo, infant rats were challenged with Hib Eagan strain, 24 h after adoptive transfer of immune sera from mice immunised with protein D-Al(OH)3. Infant rats were then bled after another 24 h and blood cultured on blood agar to measure Hib growth. The results of an experiment (Figure 7) showed that although the anti-D antibodies did not completely prevent Hib bacteraemia, it significantly reduced the mean CFU number following Hib challenge in infant rats that received protein D immune serum, compared to control animals that received non-immune serum (51 vs 122; p<0.05).
Figure 7. Protection of Infant Rats Against Hib Challenge by Protein D Antibodies
DISCUSSION
Recent data from the clinic has shown that high serum antibody titres to protein D was associated with reduced risk of developing AOM in children due to NTHi23 and studies with pneumococcal vaccines using protein D as a carrier indicated a trend of protective efficacy against AOM caused by NTHi.11,24 Moreover, Siggins et al25 demonstrated that anti-protein D antibodies led to a reduction in bacterial load in a mouse model of NTHi-lung infection but did not prevent the infection, despite high-level of protein D serum antibodies.
However, efficacy of protein D against invasive diseases caused by encapsulated Hi such as Hib has not been previously investigated and earlier research studies focused mainly on its efficacy against NTHi-induced OM. In our study, we investigated the effectiveness of protein D antibodies against Hib clinical isolates and showed that, in addition to being effective in killing Hib isolates in the bactericidal assay, protein D-induced antibodies were also protective in our in vivo infant rat Hib bacteraemia model (causing a significant 60% reduction in CFU) and this was similar to the partial protection seen in the mouse model of NTHi-lung infection described by Siggins et al.25 This suggests that full protection against Hib invasive diseases and NTHi by protein D might require the use of a more potent adjuvant26 or the combination with other highly conserved outer membrane proteins of Hi that have demonstrated potential as vaccine candidates such as P6, OMP26 and recombinant fusion protein ( protein E and Pilin A; PE-PilA).26,27,28 In addition, it is also possible that protein D could have a synergistic effect when given with Hib conjugate vaccines to improve their efficacy. However, with the high efficacy of Hib conjugate vaccines used in the infant immunisation programmes, it is difficult to measure the additional effect of protein D on Hib disease in the clinic. It remains possible, however, that including protein D in the infant immunisation, as a component vaccine or as a carrier in Synflorix®, might enable reducing the number of doses of the expensive Hib conjugate vaccine.
In this study, we investigated the effect of aluminium based-adjuvants [AlPO4 and Al(OH)3 ] and CPG on the immunogenicity of protein D and showed that all adjuvants induced a significant increase in the level of anti-protein D IgG, with Al(OH)3 inducing the highest fold increase (969) followed by AlPO4 (524) and CPG (46). It is not known how the different adjuvants potentiate the immune response, but possibly via formation of a depot at the site of immunisation and the slow release of the antigen, for the aluminium-based adjuvants29 and by activating dendritic cells through TLR9 ligation by CPG.30
Although the incidence of invasive Hib diseases declined dramatically following the introduction of Hib conjugate vaccines, infection caused by other serotypes are increasing e.g. serotype a in Alaska31,32 and serotype f in Brazil.33 Similar phenomenon of serotype replacement has been seen after the introduction of pneumococcal vaccines where clinical cases caused by serotypes not covered by the vaccine have risen whilst overall cases have fallen.34,35 Being highly conserved amongst all Hi strains, protein D is probably a good vaccine candidate that could prevent serotype replacement following the introduction of Hib conjugate vaccines and this should be investigated. Being a component of Synflorix vaccine, it would be interesting to know if Hi serotype replacement is lower in countries using Synflorix® vaccine compared to Prevenar, and the potential of protein D to target all serotypes of encapsulated Hi should be explored further.
CONCLUSION
In summary, our results showed that anti-protein D antibodies can effectively kill both Hib and NTHi strains, although not to the same degree, as it seems that the NTHi are more readily killed compared to Hib strains. We are not sure if this is due to the lower expression of protein D on the Hib strains compared to NTHi (as found in this study) or to reduced accessibility of the antibodies to bind to protein D on Hib due to the presence of the capsule and further studies are needed to explore this.
ACKNOWLEDGEMENT
The authors would like to thank the biological services team at NIBSC for excellent husbandry.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.













