1. Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981-2002. J Nat Prod. 2003; 66: 1022-1037. doi: 10.1021/np030096l
2. Berdy J. Bioactive microbial metabolites: A personal view. J Antibiot. 2005; 58: 1-26. Website. http://crawl.prod.proquest.com.s3.amazonaws.com/fpcache/e406c496973d03e7326281ef3a3966fe.pdf?AWSAccessKeyId=AKIAJF7V7KNV2KKY2NUQ&Expires=1472901756&Signature=4pJT%2BHcesaq2AouFMa%2FioR47ss0%3D. Accessed August 28, 2016
3. Pelaez F. The historical delivery of antibiotics from microbial natural products-Can history repeat? Biochem Pharmacol. 2006; 71: 981-990. doi: 10.1016/j.bcp.2005.10.010
4. Baker DD, Chu M, Oza U, Rajgarhia V. The value of natural products to future pharmaceutical discovery. Nat Prod Rep. 2007; 24: 1225-1244. doi: 10.1039/b602241n
5. Lazzarini A, Cavaletti L, Toppo G, Marinelli F. Rare genera of actinomycetes as potential producers of new antibiotics. Antonie van Leeuwenhoek. 2000; 78: 399-405. doi: 10.1023/A:1010287600557
6. Capon RJ. Microbial biodiscovery: Back to the future. Curr Top Med Chem. 2012; 12: 1471-1478. doi: 10.2174/156802612802652394
7. Clatworthy AE, Pierson E, Hung DT. Targeting virulence: A new paradigm for antimicrobial therapy. Nat Chem Biol. 2007; 3: 541-548. doi: 10.1038/nchembio.2007.24
8. Hoban DJ, Bouchillon SK, Johnson BM, Johnson JL, Dowzicky MJ. In vitro activity of tigecycline against 6792 Gramnegative and Gram-positive clinical isolates from the global Tigecycline Evaluation and Surveillance Trial (TEST Program, 2004). Diagn. Microbiol Infect Dis. 2005; 52: 215-227. doi: 10.1016/j.diagmicrobio.2005.06.001
9. Draghi DC, Tench S, Dowzicky MJ, Sahm DF. Baseline in vitro activity of tigecycline among key bacterial pathogens exhibiting multidrug resistance. Chemotherapy. 2008; 54: 91-100. doi: 10.1159/000118660
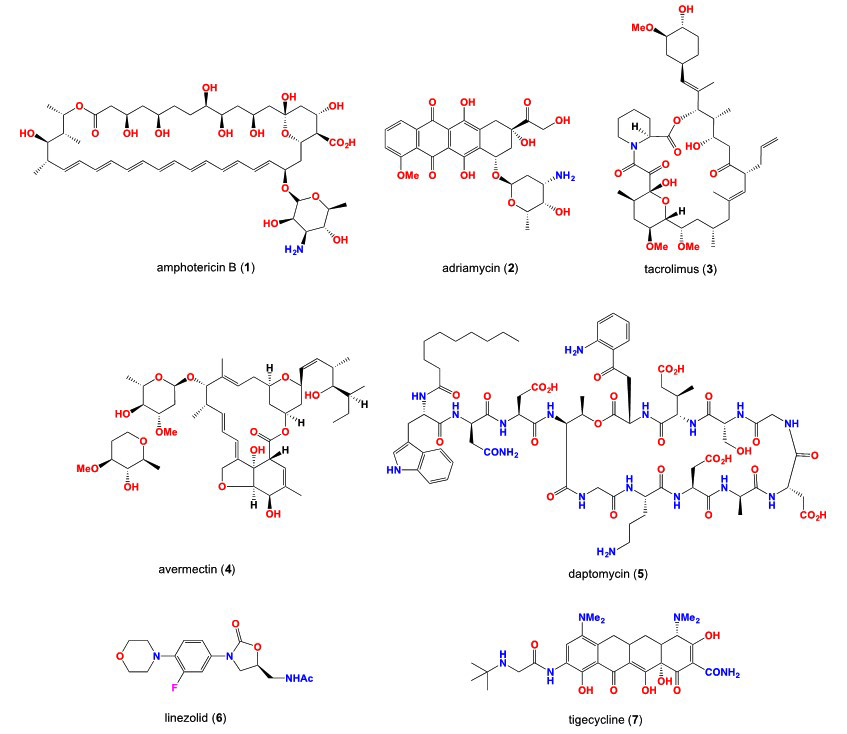
10. Wang J, Soisson SM, Young K, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006; 441: 358-361. doi: 10.1038/nature04784
11. Jayasuriya H, Herath KB, Zhang C, et al. Isolation and structure of Platencin: A FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angewandte Chemie. 2007; 46:4684-4688. doi: 10.1002/anie.200701058
12. Singh SB, Jayasuriya H, Ondeyka JG, et al. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential sensitivity strategy. J Am Chem Soc. 2006; 128: 11916-11920. doi: 10.1021/ja062232p
13. Wang J, Kodali S, Lee SH, et al. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl Acad Sci USA. 2007; 104: 7612-7616. doi: 10.1073/pnas.0700746104
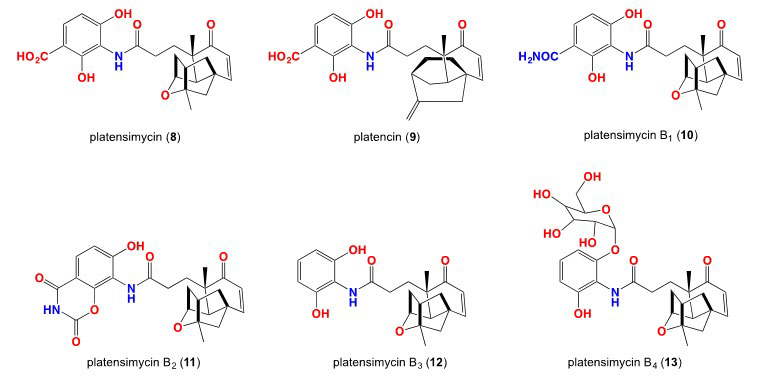
14. Zhang C, Ondeyka J, Zink DL, Burgess B, Wang J, Singh SB. Isolation, structure and fatty acid synthesis inhibitory activities of platensimycin B1-B3 from Streptomyces platensis. Chem Commun (Camb). 2008: 5034-5036. doi: 10.1039/b810113b
15. Zhang C, Ondeyka J, Guan Z, et al. Isolation, structure and biological activities of platensimycin B4 from Streptomyces platensis. J Antibiot. 2009; 62: 699-702. doi: 10.1038/ja.2009.106
16. Jacob MR, Walker LA. Natural products and antifungal drug discovery. Methods Mol Med. 2005; 118: 83-109. doi: 10.1385/1-59259-943-5:083
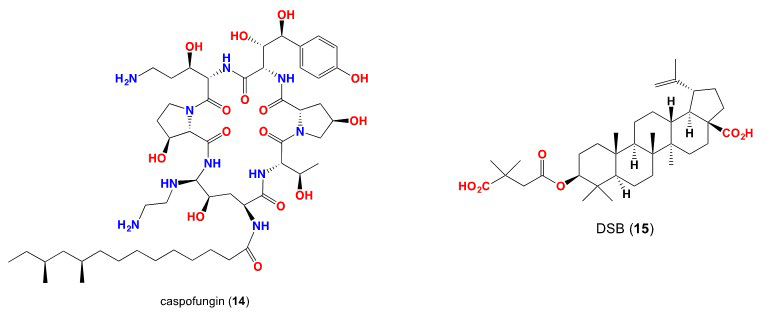
17. Deresinski SC, Stevens DA. Caspofungin. Clinical Infectious Diseases. 2003; 36: 1445-1457. doi: 10.1086/375080
18. Westby M, van Dre. CCR5 antagonists: Host-targeted antivirals for the treatment of HIV infection. Antiviral Chem Chemother. 2005; 16: 339-354. doi: 10.1177/095632020501600601
19. Ziolkowska NE, Wlodawer A. Structural studies of algal lectins with anti-HIV activity. Acta Biochim Pol. 2006; 53: 617-626. Website. http://www.actabp.pl/pdf/4_2006/617.pdf. Accessed August 29, 2016
20. Halfon P. Treatment of hepatitis C virus related diseases using hydroxychloroquine or a combination of hydroxychloroquine and an anti-viral agent. Patent Docs. 2011. Website. http://www.patentsencyclopedia.com/imgfull/20130121965_01. Accessed August 29, 2016
21. Yu D, Morris-Natschke SL, Lee K-H. New developments in natural products-based anti-AIDS research. Med Res Rev. 2007; 27: 108-132. doi: 10.1002/med.20075
22. Taubes G. The bacteria fight back. Science. 2008; 321: 356-361. doi: 10.1126/science.321.5887.356
23. Chait R, Vetsigian K, Kishony R. What counters antibiotic resistance in nature? Nat Chem Biol. 2012; 8: 2-5. doi: 10.1038/nchembio.745
24. D’Costa VM, King CE, Kalan L, et al. Antibiotic resistance is ancient. Nature. 2011; 477: 457-461. doi: 10.1038/nature10388
25. Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: Antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010; 8: 251-259. doi: 10.1038/nrmicro2312
26. Wright GD. Antibiotic resistance in the environment: A link to the clinic? Curr Opin Microbiol. 2010; 13: 589-594. doi: 10.1016/j.mib.2010.08.005
27. Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat Rev Cancer. 2002; 2: 48-58. doi: 10.1038/nrc706
28. Qin T, Skraba-Joiner SL, Khalil ZG, Johnson RP, Capon RJ, Porco JA, Jr. Atropselective syntheses of (-) and (+) rugulotrosin A utilizing point-to-axial chirality transfer. Nat Chem. 2015; 7: 234-240. doi: 10.1038/nchem.2173
29. Shang Z, Khalil Z, Li L, et al. Roseopurpurins: Chemical diversity enhanced by convergent biosynthesis and forward and reverse michael additions. Org Lett. 2016; 18: 4340-4343. doi: 10.1021/acs.orglett.6b02099
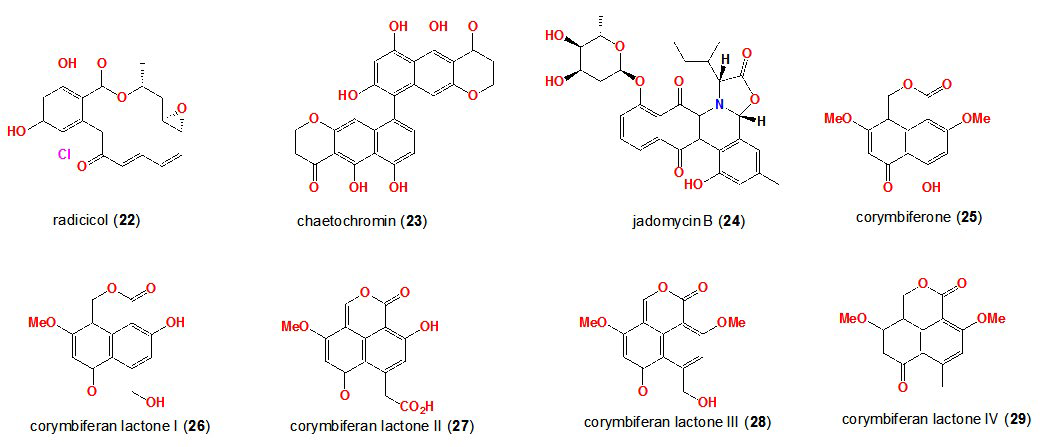
30. Shang Z, Salim AA, Khalil Z, Bernhardt PV, Capon RJ. Fungal biotransformation of tetracycline antibiotics. J Org Chem. 2016; 81: 6186-6194. doi: 10.1021/acs.joc.6b01272
31. Khalil ZG, Raju R, Piggott AM, et al. Antimycobacterial Anthracyclines from an Australian Marine-Derived Streptomyces sp. J Nat Prod. 2015; 78: 949-952. doi: 10.1021/acs.jnatprod.5b00095
32. Bok JW, Chiang Y-M, Szewczyk E, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009; 5: 462-464. doi: 10.1038/nchembio.177
33. Scherlach K, Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem. 2009; 7: 1753-1760. doi: 10.1039/b821578b
34. Omura S, Ikeda H, Ishikawa J, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001; 98: 12215-12220. doi: 10.1073/pnas.211433198
35. Bentley SD, Chater KF, Cerdeno-Tarraga AM, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002; 417: 141-147. doi: 10.1038/417141a
36. McAlpine JB, Bachmann BO, Piraee M, et al. Microbial genomics as a guide to drug discovery and structural elucidation: ECO-02301, a novel antifungal agent, as an example. J Nat Prod. 2005; 68: 493-496. doi: 10.1021/np0401664
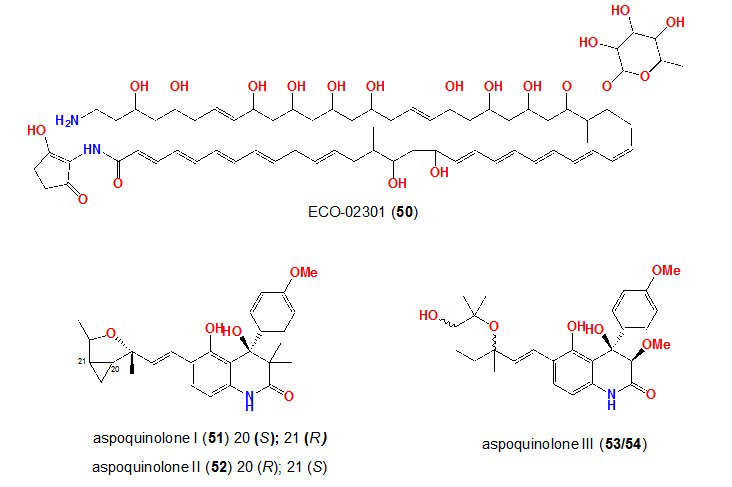
37. Udwary DW, Zeigler L, Asolkar RN, et al. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci USA. 2007; 104: 10376-10381. doi: 10.1073/pnas.0700962104
38. Elander RP. Industrial production of β-lactam antibiotics. Appl Microbiol Biotechnol. 2003; 61: 385-392. doi: 10.1007/s00253-003-1274-y
39. Paranagama PA,Wijeratne EMK, Gunatilaka AAL. Uncovering biosynthetic potential of plant-associated fungi: Effect of culture conditions on metabolite production by paraphaeosphaeria quadriseptata and chaetomium chiversii. J Nat Prod. 2007; 70: 1939-1945. doi: 10.1021/np070504b
40. Ayer SW, McInnes AG, Thibault P, et al. Jadomycin, a novel 8H-benz[b]oxazolo[3,2-f]phenanthridine antibiotic from Streptomyces venezuelae ISP5230. Tetrahedron Lett. 1991; 32: 6301-6304. doi: 10.1016/0040-4039(91)80152-V
41. Doull JL, Singh AK, Hoare M, Ayer SW. Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: Effects of heat shock, ethanol treatment and phage infection. J Ind Microbiol. 1994; 13: 120-125. doi: 10.1007/BF01584109
42. Overy DP, Smedsgaard J, Frisvad JC, Phipps RK, Thrane U. Host-derived media used as a predictor for low abundant, in planta metabolite production from necrotrophic fungi. J Appl Microbiol. 2006; 101: 1292-1300. doi: 10.1111/j.1365-2672.2006.03037.x
43. Cueto M, Jensen PR, Kauffman C, Fenical W, Lobkovsky E, Clardy J. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J Nat Prod. 2001; 64:1444-1446. doi: 10.1021/np0102713
44. Chiang Y-M, Szewczyk E, Nayak T, et al. Molecular genetic mining of the aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008; 15: 527-532. doi: 10.1016/j.chembiol.2008.05.010
45. Latifi A, Winson MK, Foglino M, et al. Multiple homologs of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995; 17: 333-343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x
46. Bode HB. No need to be pure: Mix the cultures! Chem Biol. 2006; 13: 1245-1246. doi: 10.1016/j.chembiol.2006.12.001
47. Capon RJ. Microbial biodiscovery: Back to the future. Curr Top Med Chem. 2012; 12: 1471-1478. doi: 10.2174/156802612802652394
48. Horinouchi S, Beppu T. Autoregulatory factors and communication in actinomycetes. Annu Rev Microbiol. 1992; 46: 377-398. doi: 10.1146/annurev.mi.46.100192.002113
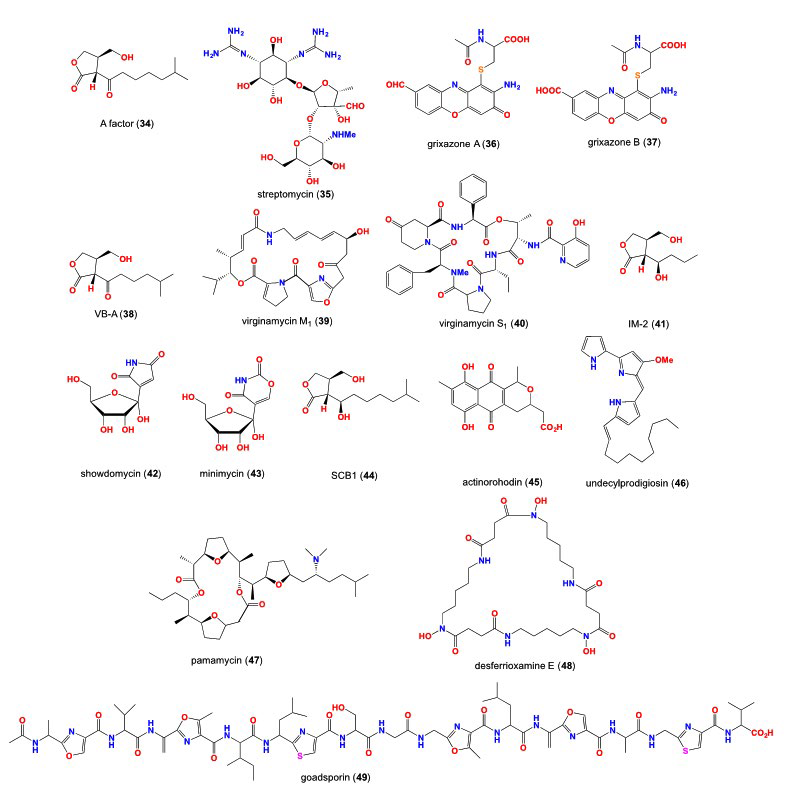
49. Kleiner EM, Pliner SA, Soifer VS, et al. Structure of the A factor, a bioregulator from Streptomyces griseus. Bioorg Khim. 1976; 2: 1142-1147.
50. Hashimoto M, Kondo T, Kozone I, Kawaide H, Abe H, Natsume M. Relationship between response to and production of the aerial mycelium-inducing substances pamamycin-607 and A-factor. Biosci Biotechnol Biochem. 2003; 67: 803-808. doi: 10.1271/bbb.67.803
51. Recio E, Colinas A, Rumbero A, Aparicio JF, Martin JF. PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J Biol Chem. 2004; 279: 41586-41593. doi: 10.1074/jbc.M402340200
52. Onaka H, Tabata H, Igarashi Y, Sato Y, Furumai T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and characterization. J Antibiot. 2001; 54: 1045-1053. doi: 10.7164/antibiotics.54.1045
53. Asad S, Opal SM. Bench-to-bedside review: Quorum sensing and the role of cell-to-cell communication during invasive bacterial infection. Crit Care. 2008; 12: 236. doi: 10.1186/cc7101
54. Antony M, Jayachandran K. Regulation of acyl homoserine lactone synthesis in pseudomonas putida jMQS1 under phenol stress. Water Air and Soil Pollution. 2016; 227: 338. doi: 10.1007/s11270-016-3018-5
55. Hwang I, Cook DM, Farrand SK. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J Bacteriol. 1995; 177: 449-458. Website. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC176609/. Accessed August 29, 2016
56. Jones B, Yu B, Bainton NJ, et al. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993; 12: 2477-2482. Website. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC413484/. Accessed August 29, 2016
57. Xie Y, Liu Z, Zhang G, et al. A rifampicin-resistant (rpoB) mutation in Pseudomonas protegens Pf-5 strain leads to improved antifungal activity and elevated production of secondary metabolites. Res Microbiol. 2016. doi: 10.1016/j.resmic.2016.05.001
58. Cheng J, Zhang X-l, Zhao J-Y, Wen M-l, Ding Z-G, Li M-G. Recent progress of study on secondary metabolites of Streptomyces. Zhongguo Kangshengsu Zazhi. 2015; 40: 791-800.
59. Sengupta S, Pramanik A, Ghosh A, Bhattacharyya M. Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem. BMC Microbiol. 2015; 15: 170. doi: 10.1186/s12866-015-0495-4
60. Zazopoulos E, Huang K, Staffa A, et al. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat Biotechnol. 2003; 21: 187-190. doi: 10.1038/nbt784
61. Scherlach K, Hertweck C. Discovery of aspoquinolones A-D, prenylated quinoline-2-one alkaloids from Aspergillus nidulans, motivated by genome mining. Org Biomol Chem. 2006; 4: 3517-3520. doi: 10.1039/B607011F
62. Perrin RM, Fedorova ND, Bok JW, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007; 3: e50. doi: 10.1371/journal.ppat.0030050
63. Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP. Genomic mining for aspergillus natural products. Chem Biol. 2006; 13: 31-37. doi: 10.1016/j.chembiol.2005.10.008
64. Shwab EK, Keller NP. Regulation of secondary metabolite production in filamentous ascomycetes. Mycol Res. 2008; 112: 225-230. doi: 10.1016/j.mycres.2007.08.021
65. Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem. 2008; 6: 1895-1897. doi: 10.1039/b804701d
66. Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002; 71: 635-700. doi: 10.1146/annurev.biochem.71.110601.135414
67. Rosenfeld Y, Shai Y. Lipopolysaccharide (Endotoxin)-host defense antibacterial peptides interactions: Role in bacterial resistance and prevention of sepsis. Biochim Biophys Acta. 2006; 1758: 1513-1522. doi: 10.1016/j.bbamem.2006.05.017
68. Khalil ZG, Kalansuriya P, Capon RJ. Lipopolysaccharide (LPS) stimulation of fungal secondary metabolism. Mycology. 2014; 5: 168-178. doi: 10.1080/21501203.2014.930530
69. Medema MH, Blin K, Cimermancic P, et al. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011; 39: W339-W346. doi: 10.1093/nar/gkr466