1. Fausto N, Campbell JS, Riehle KJ. Liver regeneration. J Hepatol. 2012; 57: 692-694. doi: 10.1002/jcp.21172
2. Jones K, Timchenko L, Timchenko NA. The role of CUGBP1 in age-dependent changes of liver functions. Ageing Research Reviews. 2012; 11: 442-449. doi: 10.1016/j.arr.2012.02.007
3. Riehle KJ, Dan YY, Campbell JS, Fausto N. New concepts in liver regeneration. J Gastroenterol Hepato.l 2011; 26 Suppl 1: 203-212. doi: 10.1111/j.1440-1746.2010.06539.x
4. Timchenko NA. Aging and liver regeneration. Trends Endocrinol Metab. 2009; 20: 171-176. doi: 10.1016/j. tem.2009.01.005
5. Michalopoulos G. Principles of liver regeneration and Growth Homeostasis. Comprehensive Physiology. 2013; 3: 485-513. doi: 10.1002/cphy.c120014
6. Michalopoulos G. Advances in liver regeneration. Expert Review of Gastroenterology. & Hepatology. 2014; 26: 1-11. doi: 10.1586/17474124.2014.934358
7. Nygard IE, Mortensen KE, Hedegaard J, et al. The genetic regulation of the terminating phase of liver regeneration. Comp Hepatol. 2012; 11: 3. doi: 10.1186/1476-5926-11-3
8. Rychtrmoc D, Hubalkova L, Viskova A, Libra A, Buncek M, Cervinkova Z. Transcriptome temporal and functional analysis of liver regeneration termination. Physiol Res. 2012; 61 Suppl 2: S77-S92.doi: 10.33549/physiolres.932393
9. Chen H, Sun Y, Dong R, et al. Mir-34a is upregulated during liver regeneration in rats and is associated with the suppression of hepatocyte proliferation. PLoS One. 2011; 6: e20238. doi: 10.1371/journal.pone.0020238
10. Yuan B, Dong R, Shi D, et al. Down-regulation of miR-23b may contribute to activation of the TGF-beta1/Smad3 signalling pathway during the termination stage of liver regeneration. FEBS Lett. 2011; 585: 927-934. doi: 10.1016/j.febslet.2011.02.031
11. Apte U, Gkretsi V, Bowen WC, et al. Enhanced liver regeneration following changes induced by hepatocyte-specific genetic ablation of integrin-linked kinase. Hepatology. 2009; 50: 844- 851. doi: 10.1002/hep.23059
12. Koral K, Paranjpe S, Bowen WC, Mars W, Luo J, Michalopoulos GK. Leukocyte specific protein-1: A novel regulator of hepatocellular proliferation and migration deleted in human HCC. Hepatology. 2014. doi: 10.1002/hep.27444
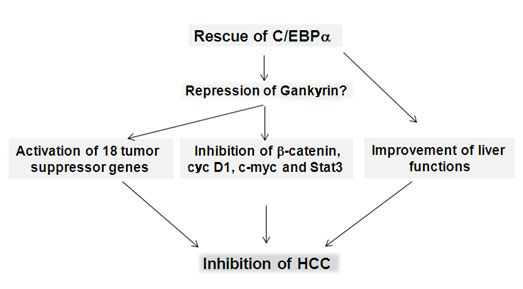
13. Jin J, Hong IH, Lewis K, et al. Cooperation of C/EBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration in mice. Hepatology. 2014. doi: 10.1002/hep.27295
14. Jin J, Wang GL, Iakova P, et al. Epigenetic changes play critical role in age-associated dysfunctions of the liver. Aging Cell. 2010; 9: 895-910. doi: 10.1111/j.1474-9726- .2010.00617.x
15. Wang GL, Shi X, Haefliger S, et al. Elimination of C/EBPalpha through the ubiquitin-proteasome system promotes the development of liver cancer in mice. J Clin Invest. 2010; 120: 2549-2562. doi: 10.1172/JCI41933
16. Hong IH, Lewis K, Iakova P, et al. Age-associated Change of C/EBP Family Proteins Causes Severe Liver Injury and Acceleration of Liver Proliferation after CCl4 Treatments. J Biol Chem. 2014; 289: 1106-1118. doi: 10.1074/jbc.M113.526780
17. Jin J, Iakova P, Breaux M, et al. Increased expression of enzymes of triglyceride synthesis is essential for the development of hepatic steatosis. Cell Rep. 2013; 3: 831-843. doi: 10.1016/j. celrep.2013.02.009
18. Jin J, Iakova P, Jiang Y, et al. Transcriptional and translational regulation of C/EBPbeta-HDAC1 protein complexes controls different levels of p53, SIRT1, and PGC1alpha proteins at the early and late stages of liver cancer. J Biol Chem. 2013; 288: 14451-14462. doi: 10.1074/jbc.M113.460840
19. Michalopoulos G. Terminating hepatocyte proliferation during liver regeneration: The roles of two members of the same family (C/EBP α and β) with opposing actions. Hepatology. 2014. doi: 10.1002/hep.27329
20. Zhao B, Wei X, Li W, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007; 21(21): 2747-2761. doi: 10.1101/gad.1602907
21. Wang C, Zhang L, He Q, et al. Differences in Yes-associated protein and mRNA levels in regenerating liver and hepatocellular carcinoma. Mol Med Rep. 2012; 5(2): 410-414. doi: 10.3892/ mmr.2011.640
22. Grijalva JL, Huizenga M, Mueller K, et al. Dynamic alterations in Hippo signaling pathway and YAP activation during liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2014; 307(2): G196-G204. doi: 10.1152/ajpgi.00077.2014
23. Yimlamai D, Christodoulou C, Galli GG, et al. Hippo pathway activity influences liver cell fate. Cell. 2014; 157(6): 1324- 1338. doi: 10.1016/j.cell.2014.03.060
24. Martin J, Dufour JF. Tumor suppressor and hepatocellular carcinoma. World J Gastroenterol. 2008; 14(11): 1720-1733. doi: 10.3748/wjg.14.1720
25. Callegari E, Gramantieri L, Domenicali M, D’Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2014. doi: 10.1038/cdd.2014.136
26. Yin H, Peng X, Ren P, Cheng B, Li S, Qin C. MicroRNAs as a novel class of diagnostic biomarkers in detection of hepatocellular carcinoma: a meta-analysis. Tumour Biol. 2014. doi: 10.1007/s13277-014-2544-2
27. Khare S, Zhang Q, Ibdah JA. Epigenetics of hepatocellular carcinoma: role of microRNA. World J Gastroenterol. 2013; 19(33): 5439-5445. doi: 10.3748/wjg.v19.i33.5439
28. Revill K, Wang T, Lachenmayer A, et al. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 2013; 145(6): 1424-35.e1-25. doi: 10.1053/j.gastro.2013.08.055
29. Xue W, Kitzing T, Roessler S, et al. SWA cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci U S A. 2012; 109(21): 8212-8217. doi: 10.1073/pnas.1206062109
30. Aguirre E, Renner O, Narlik-Grassow M, Blanco-Aparicio C. Genetic Modeling of PIM Proteins in Cancer: Proviral Tagging and Cooperation with Oncogenes, Tumor Suppressor Genes, and Carcinogens. Front Oncol. 2014; 4: 109. doi: 10.3389/ fonc.2014.00109. eCollection 2014
31. Wolosz D, Walczak A, Wilczynski GM, Szparecki G, Wilczek E, Gornicka B. Deleted in liver cancer 1 expression and localization in hepatocellular carcinoma tissue sections. Oncol Lett. 2014; 8(2): 785-788. doi: 10.3892/ol.2014.2216
32. Zimonjic DB, Popescu NC. Role of DLC1 tumor suppressor gene and MYC oncogene in pathogenesis of human hepatocellular carcinoma: potential prospects for combined targeted therapeutics. Int J Oncol. 2012; 41(2): 393-406. doi: 10.3892/ ijo.2012.1474
33. Zhou X, Thorgeirsson SS, Popescu NC. Restoration of DCL-1 gene expression induces apoptosisand inhibits both cell growth and tumorigenecity in human hepatocarcinima cells. Oncogene. 2014; 23: 1308-1313. doi: 10.1038/sj.onc.1207246
34. Zhao H, Wang J, Han Y, et al. ARID2: a new tumorsuppressor gene in hepatocellular carcinoma. Oncotarget. 2011; 2(11): 886-891.doi: 10.18632/oncotarget.355
35. Baltayiannis G, Baktayiannis N, Tsianov EV. Suppressors of cytokine signaling as tumor suppressors. Silencing of SOC3 facilitates tumor formation and growth in lung and liver. J Boun. 2008; 13: 263-265.
36. Vousden KH, Prives C. Blinded by light: the growing complexity of p53. Cell. 2009; 137: 413-431. doi: 10.1016/j. cell.2009.04.037
37. Kurinna S, Stratton SA, Coban Z, et al. p53 regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology. 2013; 57(5): 2004-2013. doi: 10.1002/ hep.26233
38. Kirstein MM, Vogel A. The pathogenesis of hepatocellular carcinoma. Dig Dis. 2014; 32(5): 545-553. doi: 10.1159/000360499
39. Hernandez-Boussard T, Rodrigez-Tome P, Montesano R, Hainaout P. IARC p53 mutation database: a rational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency fo Research on Cancer. Human Mutat. 1999; 14: 1-8. doi: 10.1002/(SICI)1098-1004- (1999)14:1<_x0031_:_x003a_AID-HUMU1>3.0.CO;2-H
40. Vaughan C, Pearsall I, Yeudall A, Deb SP, Deb S. p53: Its Mutations and Their Impact on Transcription. Subcell Biochem. 2014; 85: 71-90. doi: 10.1007/978-94-017-9211-0_4
41. Pant V, Lozano G. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev. 2014; 28(16): 1739- 1751. doi: 10.1101/gad.247452.114
42. Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000; 20(22): 8458- 8467. doi: 10.1128/MCB.20.22.8458-8467.2000
43. Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol. 2007; 27: 8284-8295. doi: 10.1128/MCB.00050-07
44. Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993; 12(2): 461- 468. doi: 10.1101/gad.1941710
45. Wu X, Bayle JH, Olson D, LevineAJ. The p53-mdm-2 auto regulatory feedback loop. Gen & Dev. 1993; 7: 1126-1132.doi: 10.1101/gad.7.7a.1126
46. Saucedo LG, Carsten BP, Seavey SE, Albee LD Perry ME. Regulation of transcriptional activity of p53 gene by p53 in response to UV radiation. Cell Growth Differ. 1998; 9: 119-130.
47. Pand V, Lozano G. Dissecting the p53-mdm2 feedback loop in vivo: uncoupling the role of p53 stability and activity. Oncotarget. 2014; 5: 1149-1156.doi: 10.18632/oncotarget.1797
48. Conner EA, Lemmer ER, Omori M, Wirth PJ, Factor VM, Thorgeirsson SS. Dual functions of E2F-1 in a transgenic mouse model of liver carcinogenesis. Oncogene. 2000 19(44): 5054- 5062.doi: 10.1038/sj.onc.1203885
49. Zhan L, Huang C, Meng XM, et al. Promising roles of mammalian E2Fs in hepatocellular carcinoma. Cell Signal. 2014; 26(5): 1075-1081. doi: 10.1016/j.cellsig.2014.01.008
50. Ghazaryan S, Sy C, Hu T, et al. Inactivation of Rb and E2f8 synergizes to trigger stressed DNA replication during erythroid terminal differentiation. Mol Cell Biol. 2014; 15: 2833-2847. doi: 10.1128/MCB.01651-13
51. Pandit SK, Westendorp B, Nantasanti S, et al. E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol. 2012; 11: 1181-1191. doi: 10.1038/ncb2585
52. Delgado I, Fresnedo O, Iglesias A, et al. A role for transcription factor E2F2 in hepatocyte proliferation and timely liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2011; 301(1): G20-G31. doi: 10.1152/ajpgi.00481.2010
53. Iakova P, Awad SS, and Timchenko NA. Aging reduces proliferative capacities of liver by switching pathways of C/ EBP growth arrest. Cell. 2003; 113: 495-506. doi: http://dx.doi. org/10.1016/S0092-8674(03)00318-0
54. Timchenko NA. Old livers: C/EBP meets new partners. Cell Cycle. 2003; 2: 445-446. doi: 10.4161/cc.2.5.467
55. Timchenko NA, Wilde M, Darlington GJ. C/EBP regulates formation of S-phase specific E2F/p107 complexes in livers of newborn mice. Mol Cell Biol. 1999; 19:2936-2945.doi: 10.1128/MCB.19.4.2936
56. Rickheim DG, Nelsen CJ, Fassett JT, Timchenko NA, Hansen LK, Albrecht JH. Differential regulation of cyclins D1 and D3 in hepatocyte proliferation. Hepatology. 2002; 36(1): 30-38. doi: 10.1053/jhep.2002.33996
57. Lee K, Lee KM, Kim TJ, et al. The nuclear 16-kD protein methylation increases in the early period of liver regeneration in a hepatectomized rat. Exp Mol Med. 2004; 36(6): 563-571. doi: 10.1038/emm.2004.72
58. Wang MJ, Chen F, Li JX, et al. Reversal of hepatocyte senescence after continuous in vivo cell proliferation. Hepatology. 2014; 60(1): 349-361. doi: 10.1002/hep.27094
59. Zhu C, Ikemoto T, Utsunomiya T, et al. Senescence-related genes possibly responsible for poor liver regeneration after hepatectomy in elderly patients. J Gastroenterol Hepatol. 2014; 29(5): 1102-1108. doi: 10.1111/jgh.12468
60. Hui AM, Makuuchi M, Li X, Cell cycle regulators and human hepatocarcinogenesis. Hepatogastroenterology. 1988; 45: 1635-1642
61. Pulling LC, Klinge DM, Belinsky SA. p16INK4a and β-catenin alterations in rat liver tumors induced by NNK. Carcinogenesis. 2001; 22(3): 461-466. doi: 10.1093/carcin/22.3.461
62. Nishida N, Kudo M. Recent advancements in comprehensive genetic analyses for human hepatocellular carcinoma. Oncology. 2013; 84 Suppl 1: 93-97. doi: 10.1159/000345897
63. Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics. 2011; 12(2): 130-137. doi: 10.2174/138920211795564359
64. Dominguez-Malagón H, Gaytan-Graham S. Hepatocellular carcinoma: an update. Ultrastruct Pathol. 2001; 25(6): 497- 516.doi: 10.1080/019131201753343539
65. Azechi H, Nishida N, Fukuda Y, et al. Disruption of the p16/ cyclin D1/retinoblastoma protein pathway in the majority of human hepatocellular carcinomas. Oncology. 2001; 60(4): 346- 354. doi: 10.1159/000058531
66. Viatour P, Ehmer U, Saddic LA, et al. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J Exp Med. 2011; 208(10): 1963-1976. doi: 10.1084/ jem.20110198
67. Ehmer U, Zmoos AF, Auerbach RK, et al. Organ size control is dominant over Rb family inactivation to restrict proliferation in vivo. Cell Rep. 2014; 8(2): 371-381. doi: 10.1016/j. celrep.2014.06.025
68. Johnson PE. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Science. 2005; 118: 2545-2455.doi: 10.1242/jcs.02459
69. Timchenko NA, Harris TE, Wilde M, et al. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol Cell Biol. 1997; 17: 7353- 7361.doi: 10.1128/mcb.17.12.7353
70. Flodby PC, Barlow H., Kalefjord L, Ahrlund-Richer L, Xanthopolous KG. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/Enhancer binding protein α. J Biol Chem. 1996; 271: 24753-24760. doi: 10.1074/ jbc.271.40.24753
71. Soriano HE, Kang DC, Finegold M, et al. Lack of C/EBPα gene expression results in increased DNA synthesis and in an increased frequency of immortalization of freshly isolated mouse hepatocytes. Hepatology. 1998; 27: 392-401.doi: 10.1002/hep.510270212
72. Wang H, Goode T, Iakova P, Albrecht J, Timchenko NA. C/ EBPα triggers proteasome-dependent degradation of cdk4 during growth arrest. EMBO J. 2002; 21: 930-941. doi: 10.1093/ emboj/21.5.930
73. Wang H, Iakova P, Wilde M, et al. C/EBPα arrests cell proliferation through direct inhibition of cdk2 and cdk4. Molecular Cell. 2001; 8: 817-828. doi: http://dx.doi.org/10.1016/S1097- 2765(01)00366-5
74. Tan EH, Hooi SC, Laban M, et al. CCAAT/Enhancer Binding Protein Knock-in Mice Exhibit Early Liver Glycogene Storage and Reduced Susceptibility to Hepatocellular Carcinoma. Cancer Res. 2005; 65: 10330-10337. doi: 10.1158/0008-5472. CAN-04-4486
75. Wang G-L, Iakova P, Wilde M, Awad S, Timchenko NA. Liver tumors escape negative control of proliferation via PI3K/ Akt-mediated block of C/EBPα growth inhibitory activity. Gen & Dev. 2004: 18:912-925. doi: 10.1101/gad.1183304
76. Wang G-L, Shi X, Salisbury E, et al. Cyclin D3 maintains growth-inhibitory activity of C/EBPα by stabilizing C/EBPαcdk2 and C/EBPα-Brm complexes. Mol Cell Biol. 2006; 26: 2570-2582. doi: 10.1128/MCB.26.7.2570-2582.2006
77. Wang G-L, Shi X, Salisbury E, Timchenko NA. Regulation of apoptotic and growth inhibitory activities of C/EBPα in different cell lines. Exp Cell Research. 2008; 314: 1626-1639. doi: 10.1016/j.yexcr.2008.01.028
78. Reebye V, Sætrom P, Mintz PJ, et al. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014; 59(1): 216-227. doi: 10.1002/ hep.26669
79. Tomizawa M, Watanabe K, Saicho H, Nakagawara A, Tagawa M. Down-regulated expression of the CCAAT/enhancer binding protein alpha and beta in human hepatocellular carcinoma: a possible prognostic marker. Anticancer Res. 2003; 23: 351-354.
80. Tseng HH, Hwang YH, Yeh KT, Chang JG, Chen YL, Yu HS. Reduced expression of C/EBPα protein in hepatocellular carcinoma is associated with advanced tumor stage and shortened patient survival. J Cancer Res Clin Oncol. 2009; 135: 241-247. doi: 10.1007/s00432-008-0448-5
81. Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J Gastroenterol. 2014; 20(1): 22-30. doi: 10.3748/wjg.v20.i1.22
82. Parviz F, Matullo C, Garrison WD, et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003; 34(3): 292-296. doi: 10.1038/ng1175
83. Lemaigre F, Zaret KS. Liver development update: new embryo models, cell lineage control, and morphogenesis. Curr Opin Genet Dev. 2004; 14(5): 582-950. doi: 10.1016/j. gde.2004.08.004
84. Chellappa K, Jankova L, Schnabl JM, et al. Src tyrosine kinase phosphorylation of nuclear receptor HNF4α correlates with isoform-specific loss of HNF4α in human colon cancer. Proc Natl Acad Sci U S A. 2012; 109(7): 2302-2307. doi: 10.1073/ pnas.1106799109
85. Yao D, Peng S, Dai C. The role of hepatocyte nuclear factor 4alpha in metastatic tumor formation of hepatocellular carcinoma and its close relationship with the mesenchymalepithelial transition markers. BMC Cancer. 2013; 13: 432. doi: 10.1186/1471-2407-13-432
86. Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014; 513(7518): 382-387. doi: 10.1038/nature13438
87. Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J Biol Chem. 2012; 287(10): 7345-7356. doi: 10.1074/jbc.M111.334599
88. Hatziapostolou M, Polytarchou C, Aggelidou E, et al. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011; 147(6): 1233-1247. doi: 10.1016/j.cell.2011.10.043
89. Ning BF, Ding J, Yin C, et al. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res. 2010; 70(19): 7640-7651. doi: 10.1158/0008-5472. CAN-10-0824
90. Walesky C, Gunewardena S, Terwilliger EF, et al. Hepatocyte-specific deletion of hepatocyte nuclear factor-4α in adult mice results in increased hepatocyte proliferation. Am J Physiol Gastrointest Liver Physiol. 2013; 304(1): G26-37. doi: 10.1152/ ajpgi.00064.2012
91. Walesky C, Edwards G, Borude P, et al. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology. 2013; 57(6): 2480-2490. doi: 10.1002/hep.26251
92. Yang M, Li SN, Anjum KM, et al. A double-negative feedback loop between Wnt-β-catenin signaling and HNF4α regulates epithelial-mesenchymal transition in hepatocellular carcinoma. J Cell Sci. 2013; 126(Pt 24): 5692-5703. doi: 10.1242/ jcs.135053
93. Alder O, Cullum R, Lee S, et al. Hippo Signaling Influences HNF4A and FOXA2 Enhancer Switching during Hepatocyte Differentiation. Cell Rep. 2014 Sep 24. pii: S2211-1247- (14):00722-00730. doi: 10.1016/j.celrep.2014.08.046
94. Saha SK, Parachoniak CA, Ghanta KS, et al. Mutant IDH inhibits HNF-4α to block hepatocyte differentiation and promote biliary cancer. Nature. 2014; 513(7516): 110-114. doi: 10.1038/ nature13441
95. Dawson S. Hepatocellular carcinoma and ubiquitin-proteasome system. Biochem. Biophys Acta. 2008; 1782: 775-784. doi: 10.1016/j.bbadis.2008.08.003
96. Lim IK. Spectrum of molecular changes during hepatocvarcinogenesis induced by DEN and other chemical in Fishes 344 rats. Mech. Ageing Dev. 2003; 124: 679-708.doi: 10.1016/s0047-6374(02)00087-8
97. Krzywda S, Brzozowski AM, Higashitsuj H, et al. The crystal structure of Gankyrin, an oncoprotein found in complexes with cyclin-dependent kinase 4, a 19 S proteasomal ATPase regulator, and the tumor suppressors Rb and p53. J Biol Chem. 2004; 279(2): 1541-1545. doi: 10.1074/jbc.M310265200
98. Fu HY, Wang HY, Tan L, Liu SQ, Cao HA, Wu MC. Overexpression of p28/Gankyrin in human hepatocellular carcinoma and its clinical significance. World J. Gastroenterol. 2002; 8: 638-643.doi: 10.3748/wjg.v8.i4.638
99. Jing H, Zhang G, Meng L, Meng Q, Mo H, Tai Y.Gradually elevated expression of Gankyrin during human hepatocarcinogenesis and its clinicopathological significance. Sci Rep. 2014; 4: 5503. doi: 10.1038/srep05503
100. Li H, Fu X, Chen Y, et al. Use of adenovirus-delivered siRNA to target oncoprotein p28GANK in hepatocellular carcinoma. Gastroenterology. 2005; 128: 2029–2041. doi: http://dx.doi.org/10.1053/j.gastro.2005.03.001
101. Song X, Wang J, Zheng T, et al. LBH589 Inhibits proliferation and metastasis of hepatocellular carcinoma via inhibition of Gankyrin/STAT3/Akt pathway. Mol Cancer. 2013; 12(1): 114. doi: 10.1186/1476-4598-12-114
102. Li J, Tian F, Li D, Chen J, Jiang P, Zheng S, Li X, Wang S. MiR-605 represses PSMD10/Gankyrin and inhibits intrahepatic cholangiocarcinoma cell progression. FEBS Lett. 2014; 588(18): 3491-3500. doi: 10.1016/j.febslet.2014.08.008
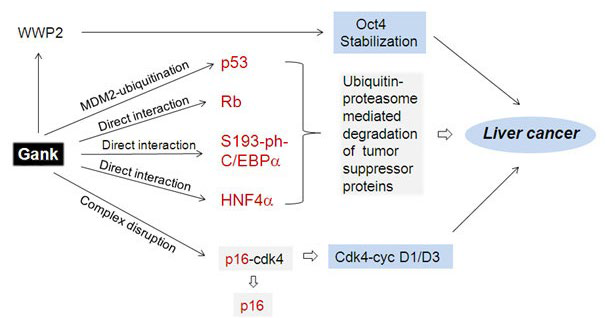
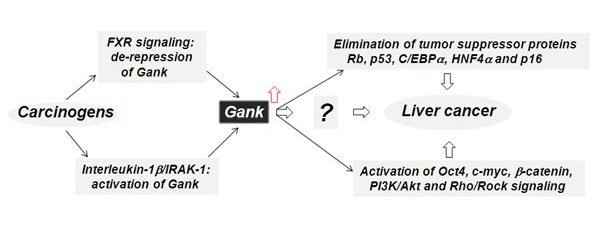
103. Jiang Y, Iakova P, Jin J, et al. Farnesoid X receptor inhibits Gankyrin in mouse livers and prevents development of liver cancer. Hepatology. 2013; 57: 1098-1106. doi: 10.1002/hep.26146
104. Higashitsuji H, Itoh K, Sakurai T, et al. The oncoprotein Gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005; 8: 75-87. doi: http://dx.doi.org/10.1016/j.ccr.2005.06.006
105. Higashitsuji H, Itoh K, Nagao T, et al. Reduced stability of retinoblastoma protein by Gankyrin, an oncogenic ankyrinrepeat protein overexpressed in hepatomas. Nature Medicine. 2000; 6: 96-99.doi: 10.1038/71600
106. Li J, Tsai MD. Novel insights into the INK4-CDK4/6-Rb pathway: counter action of Gankyrin against INK4 proteins regulates the CDK4-mediated phosphorylation of Rb. Biochemistry. 2002; 41: 3977-3983. doi: 10.1021/bi011550s
107. Sun W, Ding J, Wu K, et al. Gankyrin-mediated dedifferentiation facilitates the tumorigenicity of rat hepatocytes and hepatoma cells. Hepatology. 2011; 54(4): 1259-1572. doi: 10.1002/ hep.24530
108. Qian YW, Chen Y, Yang W, et al. p28(GANK) prevents degradation of Oct4 and promotes expansion of tumor-initiating cells in hepatocarcinogenesis. Gastroenterology. 2012; 142: 1547-1558. doi: 10.1053/j.gastro.2012.02.042
109. Zheng T, Hong X, Wang J, et al. Gankyrin promotes tumor growth and metastasis through activation of IL-6/STAT3 signaling in human cholangiocarcinoma. Hepatology. 2014; 59(3): 935-946. doi: 10.1002/hep.26705
110. Bai Z, Tai Y, Li W, et al. Gankyrin activates IL-8 to promote hepatic metastasis of colorectal cancer. Cancer Res. 2013; 73(14): 4548-4558. doi: 10.1158/0008-5472.CAN-12-4586
111. Dong LW, Yang GZ, Pan YF, et al. The oncoprotein p28GANK establishes a positive feedback loop in β-catenin signaling. Cell Res. 2011; 21(8): 1248-1261. doi: 10.1038/ cr.2011.103
112. Fu J, Chen Y, Cao J, et al. p28GANK overexpression accelerates hepatocellular carcinoma invasiveness and metastasis via phosphoinositol 3-kinase/AKT/hypoxia-inducible factor1α pathways. Hepatology. 2011; 53(1): 181-192. doi: 10.1002/ hep.24015
113. Man JH, Liang B, Gu YX, et al. Gankyrin plays an essential role in Ras-induced tumorigenesis through regulation of the RhoA/ROCK pathway in mammalian cells. J Clin Invest. 2010; 120(8): 2829-2841. doi: 10.1172/JCI42542
114. Mine H, Sakurai T, Kashida H, et al. Association of Gankyrin and stemness factor expression in human colorectal cancer. Dig Dis Sci. 2013; 58(8): 2337-2344. doi: 10.1007/ s10620-013-2627-8
115. Su B, Luo T, Zhu J, et al. Interleukin-1β/IRAK-1 inflammatory signaling contributes to persistent Gankyrin activation during hepatocarcinogenesis. Hepatology. 2014. doi: 10.1002/ hep.27551