INTRODUCTION
The Lactobacillus is a species of bacteria with many different types. These probiotic bacteria routinely live in our body systems without causing disease. Lactobacillus, which is used for the treatment of diarrhea, can be found in some fermented foods, including cheese, beer, yoghurt, cocoa, and animal feed. The Lactobacillus is a genus of Gram-positive, non-sporing, non-respiring cocci or rods, and can form spirals or spheres under certain conditions. All their energy is obtained by converting glucose into lactic acid during the fermentation of pure lactic acid. The Lactobacillus bacteria are commonly used as probiotic supplements in the dairy industry and starter cultures. There are many strains of lactic acid bacteria and are commonly used to process and ferment food.1 The consumers’ intake contains probiotics mainly in the form of dietary foods and supplements. They can be used as complementary and alternative medicine (CAM). The Lactobacillus bacteria, non-lactic acid bacteria and yeasts can be considered as probiotics. The Lactobacillus bacteria are the most important probiotic known to have beneficial effects on the human gastro-intestinal (GI) tract. Moreover, dark chocolate, microalgae, kimchi, tempeh and kombucha tea are also sources of Lactobacillus bacteria.2
A number of health benefits have been claimed from probiotic bacteria such as Lactobacillus acidophilus, Bifidobacterium spp., and Lactobacillus casei. Because of the potential health benefits, these organisms are increasingly incorporated into dairy foods.3
The Lactobacillus bacteria have a positive effect on health when its concentration is high enough. These foods can have a positive effect on the digestion and can prevent unwanted bacteria from spreading infections or diseases. Consumers are very well aware of the fact that fermented products contain living organisms, provided the fact that dairy products such as curd have a healthy record.4
Some studies have shown that some provocative cholesterol levels can be reduced with the aid of these probiotic bacteria. In addition, invasive plants and probiotics such as Lactobacillus acidophilus can help prevent and reduce diarrhea associated with various diseases. It also can help to treat and prevent vaginal infections, cold and flu symptoms, and allergy symptoms.3
Antibiotic resistance has become a serious problem due to the high number of antibiotic-resistant strains. Once the factors related to drug resistance are transferred to other microorganisms, especially through food transporters, they can cause enormous problems. The evolution of antibiotic-resistant pathogens being resistant to antibiotics has been widely reported.5
The antibiotic susceptibility of the tested strains was evaluated according to the antimicrobial drug sensitivity standard of Clinical and Laboratory Standards Institute (CLSL) criteria. A study which used 29 isolated strains of Lactobacillus, reported antibiotic resistance in the 1980s as well. The researchers generally believed that the resistance was a result of the long evolution and it was generally endogenous resistance.6
Antibiotics remain to be the main treatment strategy for the treatment of a variety of infectious pathogens in humans and veterinary medicine. However, the indiscrimination and improper use of antibiotics has led to decreased susceptibility and increased resistance rates observed not only in disease-causing microbes but in commensal microbes as well. In the human clinical environment, these antibiotic-resistant pathogens have caused numerous therapeutic failures, eventually leading to hospital morbidity and death. Many studies have shown that microbes are commonly used to produce municipal and sometimes mutated microorganisms, rather than killing pathogenic microorganisms with antibiotics.7 Probiotic bacterial strains used in both animal and human applications also have risks in becoming conduits themselves in spreading antibiotic resistance genes.8
Lactobacillus bacteria have also been used to treat diabetes, abdominal muscle syndrome, Crohn’s disease and large intestine stroke. Moreover, there are some very important benefits of the Lactobacillus bacteria such as levels of stimulating cholesterol can be reduced. In addition to that, invasive plants and probiotics like the Lactobacillus acidophilus can help prevent diarrhea related to various diseases. Most of the commercial meals and food are now associated with a set of antibiotic bacteria to meet healthy growth satisfaction requirements.9,10
However, this study is most importantly carried out to identify the probiotic bacteria Lactobacillus based on their phenotypic features and genotypic features. This study also shows the importance of probiotic bacterium, and the effects of their antibiotic resistance to human.
MATERIALS AND METHODOLOGIES
Bacterial Medium and Sample Preparation
Initially, Lactobacilli man, rogosa and sharpe (MRS) agar medium was prepared and it was autoclaved. In the meantime, 6 plates were labeled from sample A-F. The mixture was poured to the plates and it was kept to solidify by leaving it open for 30-seconds before closed. Six different curd brands were used and it was transferred to fresh beakers. After transferring curd samples were dissolved in saline water and mixed well with the glass rod.
Culturing of Lactobacillus
The sterilized loop was used to remove a loopful of inoculum from the plate. The lid of the plate was slightly lifted just enough to insert the loop. One plate was used to culture one sample. Therefore, each plate was labeled with necessary information. The inoculum was streaked by gently moving the loop in a zig-zag pattern on the agar and the plates were then placed in the incubator for 24-hours at 37 °C.
DNA Extraction
Boiled cell method:
Following standard aseptic conditions, 500 μl of autoclaved distilled water was added to each falcon tube followed by the inoculation of sufficient amount of sample into the water. The falcon tubes were centrifuged at 4000 rpm for 10-minutes and it was repeated thrice. After that the supernatant was removed. Then 500 μl of Tris-EDTA (TE) buffer was added to the tubes and it was vortexed briefly. Thereafter, the tubes were heated 100 °C for 20 °C minutes and immediately the tubes were cooled at -20 °C for 20-minutes. Then the tubes were centrifuged at 13000 rpm for 3-minutes. Finally, the falcon tubes were transferred to the 1.5 μl eppendorf tubes and stored at -5 °C freezer.
CTAB Extraction
At first 5 auto-claved falcons were labeled (Sample A-F) and 2.5 ml of autoclaved cetyl trimethylammonium bromide (CTAB) buffer were added to each falcon tubes. Then the sample was picked in sterilized loop and it was inoculated in CTAB buffer. Thereafter it was centrifuged at 4000 rpm for 10-minutes and it was repeated thrice following to the incubation of 65 °C for 30- min. Then supernatant was transferred to the fresh falcon tube and equal amounts of isopropanol alcohol were added and tube was inverted to mix. Thereafter, the tubes were centrifuged until the precipitate was observed. After receiving the pellet, the supernatant was removed without disturbing the pellet. After removing the supernatant, 2.5 ml of 70% ethanol was added to wash the pellet and it was centrifuged for 1-minute. Then the excess ethanol was taken out using micropipette and the pellet was dried for 24-hours.
Finally, 500 μl of TE was added to dissolve the pellet. The same procedure was repeated for each 6 tubes and stored at -18 °C.
Spectrophotometric DNA Quantification
Deoxyribonucleic acid (DNA) quantification for the two extraction methods was performed in a spectrophotometer to calculate the DNA concentration and DNA yield for each sample.
Readings were taken at absorbance wave length of 230 nm, 260 nm and 280 nm. These readings were taken for each of the 6 samples (Sample A-F). Before obtaining the readings the spectrophotometer was blanked using the TE cuvette. Three readings for each absorbance were taken and the mean was calculated.
• DNA concentration=260 nm (OD)×50 ug/mL×Dilution factor (DF) (101)
• DNA yield=DNA concentration×Volume of the sample (30)
After the calculation of the DNA concentration and yield, the purity of the DNA (260/280) was calculated for both extraction methods and compared.
PCR-based Identification of Lactobacillus
A seven 500 μl eppendorf tubes were placed for each reaction, and labeled appropriately to set up the number of reactions with negative control reaction. Genus specific primers were used for detection of Lactobacillus in the samples. The primers sequence are shown in below (Table 1).
| Table 1. Lactobacillus Forward and Reverse Primer Sequences |
|
Target Organism
|
Primer Set |
Sequence (5’ to 3’) |
Product Size (bp) |
Ta (°c), time (S)
|
| Lactobacillus genus |
Lacto-16S-F |
GGA ATC TTC CAC AAT GGA CG |
216
|
56, 10 S
|
| Lacto-16S-R |
CGC TTT ACG CCC AAT AAA TCC GG |
75%
|
|
The required volume was calculated for each of the polymerase chain reaction (PCR) master mix (Table 1). From the PCR master mix 12.25 μl was added into each PCR tube. Finally the DNA was added.
Detection and Analysis by Agarose Gel Electrophoresis
The following procedure was followed for all respective tests which are performed in this report after PCR. Initially, 2% agarose gel was prepared, and 8 μl of each samples and negative control was loaded onto the wells followed by 2 μl of the DNA ladder (100 bp). Then the gel was run at 45 V for 20-minutes followed by 55 V for 45-minutes.
Antibiotic Resistance Determination Using Disk Diffusion Method
To prepare the bacterial inoculum, 0.85% saline was prepared and the colony used to inoculate broth was scraped and suspended in the saline. Once the optical density matched that of the 0.5 McFarland standard, using spread plate method, 200 μl of inoculum was spread onto Muellerhinton agar. Thereafter, 10 mg discs of tetracycline, erythromycin, ampicillin, vancomycin and a negative control filter paper containing autoclaved distilled water was placed onto the agar. The plates were then incubated at 37 °C for 24-hours.
PCR Detection of Resistance Genes
PCR was performed with erythromycin erm(B) and tetracycline tet(M) specific primers and antibiotic resistance genes corresponding to the samples represented the positive results for Lactobacillus.
RESULTS
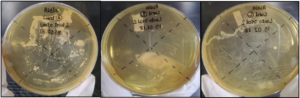
As shown in Figure 1, dull white colonies with non-defined borders were observed in sample C.
Figure 1. Curd Samples A, B and C
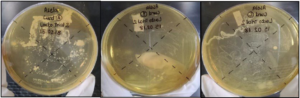
As shown in Figure 2, the colonies observed in sample D were white color rounded multiple colonies, with non-defined boarders. The colonies observed in sample E were white, irregular in shape and flat with defined borders. Multiple colonies along with various sizes were observed in sample F, which were white, round and flat with well-defined borders.
Figure 2. Curd Samples D, E and F. The Colonies Observed in Sample D were White Color Rounded Multiple Colonies, with Non-defined Boarders
Statistical Analysis
Tests of between-subjects effects:
Dependent Variable: DNA Yield The assumed hypothesis was that there is a significance between the DNA yield, the brand and method of extraction. Taking the statistical calculations into consideration as given in Table 2, the method used had a p value greater than 0.05 (0.513) and Brand had a p value less than 0.05 (0.000), respectively indicating that there was no significance between the DNA yield and the method used. But considering both the method and brand together (Method*Brand), the p value was 0.000 (<0.05) representing that there is a significant contribution from both method and brand to the yield of DNA.
| Table 2. The DNA Yield Comparisons Between Two Methods and Six Brands |
|
Source
|
df |
f value |
p value
|
| Corrected Model |
11
|
111.962 |
0.000
|
| Intercept |
1
|
1347.338 |
0.000
|
| Method |
1
|
0.441 |
0.513
|
| Brand |
5
|
109.716 |
0.000
|
| Method*Brand |
5
|
136.513 |
0.000
|
| Error |
24
|
|
|
| Total |
36
|
|
|
| Corrected Total |
35
|
|
|
| a. R Squared=0.981 (Adjusted R Squared=0.972) |
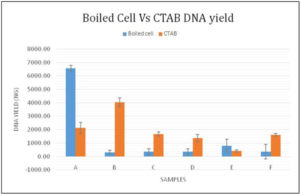
As shown by Figure 4, boiled cell method samples of A and E represented with higher DNA yielding. The samples of B, C, D and F represented higher DNA yielding by the CTAB extraction method. Comparatively CTAB extraction method’s average DNA yielding is relatively higher than the boiled cell method.
Figure 4. Graphical Distribution of DNA Yield Between Two Extraction Methods for the Sample A to F
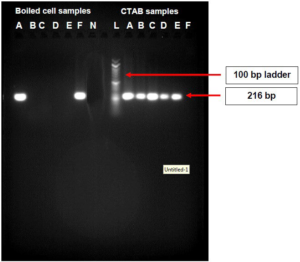
As shown in Figure 5, the boiled cell method produced bands for samples A and, whereas the CTAB method produced bands for samples A, B, C, D and E with molecular weight of 216 bp,
Figure 5. Agarose Gel Image of PCR Products of the DNA Extracted from CTAB and Boiled Cell Method
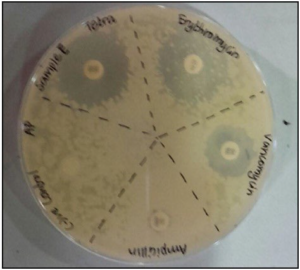
Sample A demonstrated a zone of inhibition and resistance to antibiotics of erythromycin, tetracycline and vancomycin and did not indicate any resistance to ampicillin, as shown in Figure 6.
Figure 6. The Inhibition Zones on Mueller Hintons Agar Plate of Sample A
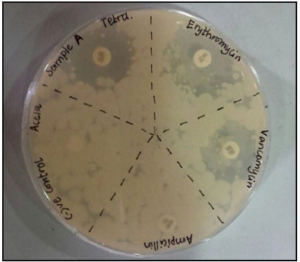
Figure 7 shows overlapping of the inhibition zones of all the antibiotics indicating the sample B being resistant to all the antibiotics.
Figure 7. The Inhibition Zones on Mueller Hintons Agar Plate of Sample B
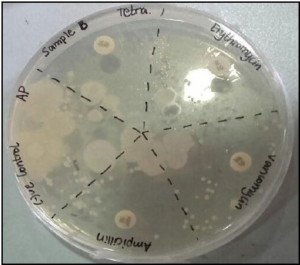
As shown by Figure 8, sample C exhibits resistance to antibiotics represented by the inhibition zones being overlapped.
Figure 8. The Inhibition Zones on Mueller Hintons Agar Plate of Sample C
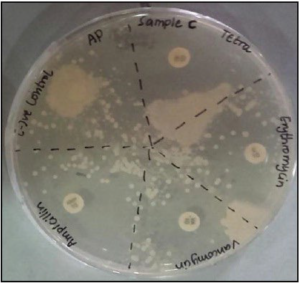
As shown in Figure 9, sample D was resistant to the antibiotic erythromycin, tetracycline and vancomycin but did not indicate any resistance to ampicillin.
Figure 9. The Inhibition Zones on Mueller Hintons Agar Plate of Sample D
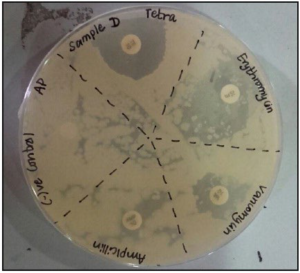
Figure 10 demonstrated the sample D being resistant to the antibiotic erythromycin, tetracycline and vancomycin but did not indicate any resistance to ampicillin.
Figure 10. The Inhibition Zones on Mueller Hintons Agar Plate of Sample E
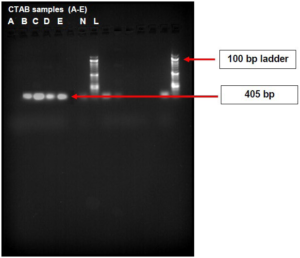
As presented in Figure 11, the erm(B) antibiotic resistance gene was detected in samples B, C, D, & E, except sample A which indicated the positive results by PCR.
Figure 11. Agarose Gel Image of PCR Products of the Positive DNA Samples Tested (CTAB) for Erythromycin Resistance Gene erm(B)
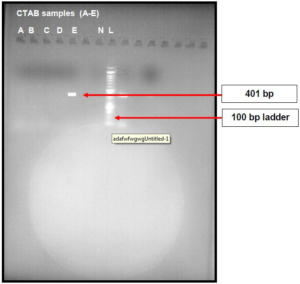
As shown in Figure 12, the tet(M) antibiotic resistance gene was only detected in sample E. The samples of B, C & D which indicated positive results in PCR did not generate any bands.
Figure 12. Agarose Gel Image of PCR Products of the Positive DNA Samples Tested (CTAB) for Tetracycline Resistance Gene tet(M).
DISCUSSION
The aim of the current study was to identify probiotic bacteria in commercially available food products and to analyze their antibiotic resistance. Generally, the lactic acid bacteria show delayed growth and smaller colony size than other microorganisms, making it impossible to differentiate each strain. However, the colony characterization was carried out and they were found to be Lactobacillus species but, various lactic acid baths (LABs) produced colonies which were quite identical, making it difficult to differentiate each species.11 MRS agar encourages the growth of the lactic acid bacteria including the species of: Lactobacillus, Streptococcus, Pediococcus and Leuconostoc genera which can produce lactic acid in considerable amounts, as shown in Figures 1 and 2. There is no medium yet available to particularly culture only the Lactobacilli. The isolated bacteria were observed through compound microscope. As shown in Figure 3, the gram staining results of all six samples indicated different types of morphologies and characteristics. Within the rod-shaped bacteria, different groups such as the diplobacillus and streptobacillus were also identified (such as Bacillus cereus).12 There were Pleomorphic, who have one or more forms depending on the phase of the cell cycle during the bacterial cell growth. In addition to the strains, variability can occur as a result of different growth conditions (medium composition, temperature and pH status). In that case the isolated bacteria could be assumed as Lactobacillus acidophillus, Lactobacillus bulgaricus, Lactobacillus mali or Lactobacillus yamanashiensis. 13,14 The Gramnegative bacteria identified as pink, short plump rods were assumed to be coliform bacterium. The coliform bacterium seems to survive despite inhibition and/or inactivation by the lactic acid bacteria present in the sample. Survival of coliform bacterium in fermented dairy product depend on variable factors such as, on the species of the lactic acid bacteria that was used to prepare the product; strains of sub-species level of inoculums used; incubation temperature at which fermentation is carried out; amount and speed of acid production; resulting pH; temperature at which the product is stored after fermentation; and composition of the product. There is a high possibility that undesirable microorganisms including pathogenic ones may be introduced into the milk through earthen pot, food handlers, starter culture, etc. Coliform bacteria have a long history of being used as microbial hygiene indicators in the U.S. dairy industry, dating back to 1914.15 A study conducted by Hervet and colleagues showed that a wide range of dairy relevant G-bacteria indicated post pasteurization contamination and other hygiene issues, which can go undetected on coliform selective and differential media.16 The Lactobacilli was the fact that they were unable to produce endospores. Many times, endospores are seen in the Gram-stain if they are present. None were observed in the Gram stain. To double check staining technique and to eliminate error, the bacteria should also be plated on MacConkey’s agar. The bacterial DNA were extracted out via two different methods which were the boiled cell method and CTAB (Cetyl-trimethyl ammonium bromide) method. The quality and yield of the extracted DNA were evaluated using UV visible spectrophotometer at 260 nm and 280 nm. The quality of DNA was determined by A260/A280 ratio value. The DNA yield, in terms of DNA concentration was calculated. In boiled cell method, sample A & sample F contained relatively high DNA concentration compared with CTAB method samples of A and F. The DNA concentration of samples B, C, D, E in boiled cell method represented relatively low DNA concentration compared with the CTAB method samples of B, C, D, E. The overall beneficial average DNA concentrations were obtained by the CTAB extraction method. As shown in Figure 4, a comparably quality DNA was obtained by boiling cell method from sample A and F. The rest of the boiled cell method samples (B, C, D, E) indicated the contamination of protein. In CTAB method total samples signified contamination of protein. The DNA concentration of each sample was calculated by obtaining absorbance at 260 nm by spectrophotometry analysis. The CTAB method extracted DNA samples were implicated to polymerase chain reaction (PCR). The identification of Lactobacillus genus was performed by genus specific PCR. The sequences of the primers of, 5’-CTC AAA ACT AAA CAA AGT TTC-3’ was used as the forward primer and 5’-CTT GTA CAC ACC GCC CGT CA-3’ was used as the reverse primer.17 Agarose gel electrophoresis was conducted for both boiled cell method and CTAB DNA extraction methods. In boiled cell method samples of A and F represented with bands. In CTAB method, samples A, B, C, D, E except F represented with bands. In sample D, a majority of G-short, plump rod bacteria were observed along with G+ rods in chains. The PCR technique is highly sensitivity and specificity because of the use of genus specific primers for amplification, which in turn increases the probability of amplification of any single genus Lactobacillus by the genus specific primers (Lacto-F/Lacto-R), that are complementary to the DNA region targeted for amplification under specific thermal cycling conditions. All the Lactobacillus strains used in this study were positive to the Lactobacillus genus specific primer set while other bacteria proved to be negative. Therefore, this primer set proved to be highly specific even when it was used for DNA extracted from complex microbial communities (CMC), which is full of mixed bacteria, fragmented DNA and PCR suppressor proteins. Under the described PCR conditions, the five different samples of Lactobacillus spp. generated the expected PCR product at molecular weight of 216 bp (base pair). The PCR assays need to be optimized and standardized prior to any valid testing, especially to detect microflora in highly mixed complex environments, CMC.18 Dairy processors across the world test for the presence of different gram-negative bacteria (Enterobacteriaceae family & coliform group) and hygiene indicators to assess the quality of their finished products. They evaluate sanitation practices at the processing-level and detect instances of post-pasteurization contaminations.16
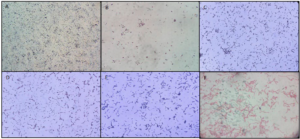
Figure 3.Sample A Shows Concentrated Gram-positive Small Rod Shaped Micr oorganism; Sample B Shows Less Concentrated Gram Positive, Slender Bacilli Rods; Sample C Shows Moderately Concentrated Gram Positive Short and Long Rods of Bacilli; Sample D Shows Gram Negative Short Plump Rod Bacteria Along with Some Gram Positive Rods in Chain Palisade Forms; Sample E Shows Moderately Concentrated Gram Positive Long, Slender rod Shaped Bacilli; and Sample F Shows Gram Negative Long Rods and Coccoid forms of Bacteria
Antibiotic susceptibility was evaluated by disk diffusion and PCR based resistance genes tet(M) and erm(B). In disk diffusion method, samples of A, D and E demonstrated resistance to the antibiotic erythromycin, tetracycline and vancomycin. These samples did not indicate any resistance to ampicillin. Samples B & C did show some resistance to antibiotics but it was difficult to determine to which antibiotics they were resistant, based on the inhibitory zones. It was observed that certain antibiotics were highly resisted by Lactobacillus. It can be assumed that it is due to high resistance that large inhibitory zones overlapped in the same plate. The range of the diameter of the zones of all antibiotics against the isolates were given. Inhibition-zone diameters were measured in millimeters in which a diameter of zone >20 mm was referred as sensitive while a diameter ≤14 mm was considered as resistant.19 Both tet(M) & erm(B) antibiotic resistance genes were detected in sample E. The samples B, C and D had resistant genes to erm(B).
CONCLUSION
This study demonstrated a wide range of resistance among Lactobacillus isolates. The increased use of fermented food products and probiotics as food supplements and healthpromoting products, contain massive amounts of bacteria acting as either donors and/or recipients of antibiotic resistant genes in the human GI tract. They also contribute to the emergence of antibiotic-resistant strains. From a safety point of view, when a bacterial strain demonstrates the resistance to antimicrobials by phenotypic methods, it is enviable to monitor the molecular basis of this resistance. In this regard, the performance of antimicrobial susceptibility testing might be considered as an essential selection criterion for probiotic cultures and an effective guide for specific antimicrobial therapy. Subsequently, strict quality control measures and the proper characterization and maintenance of starter culture strains during the production of fermented and pharmaceutical products are mandatory.
FURTHER WORK
1. The conventional culture-based methods are time consuming. Hence a new technique of immuno-magnetic separation can be applied to increase the specificity of the culture method. This technique specifically separates the target organism from other organisms. This thereby results in a useful sample for PCR with little or no nonspecific DNA and interfering factors.
2. Since the conventional microbial detection method consumes large amount of time (more than 48 hours) to identify the organism. The present study can be focused on using new and alternative molecular methods such as biosensor-based methods for detecting low concentrations of the organisms in a short amount of time. These methods have high sensitivity and high specificity.
3. Apart from microscopic observation all the samples are subjected to the biochemical tests on the basis of carbohydrate fermentation test, motility test, catalase and oxidase test to identify the bacterial isolates.
4. The gram staining procedure should be conducted by taking multiple colonies from the same sample or conducted by preparing multiple slides in the same sample for microscopic analysis.
5. For purifying DNA/RNA binding proteins, purification steps should implicate to obtain the quality DNA.
6. The PCR assays need an optimization and standardization prior to any valid testing, especially for detecting microflora in highly mixed complex environments, complex microbial communities.
7. Multiplex PCR (mPCR) is a rapid technique utilized in the rapid determination, in contrast with sole PCR, by the instant amplification of many genetic factor targets. Multiplex PCR can distinguish up to five or more microorganisms parallel. mPCR will assist in the identification of numerous species, together with Lactobacillus spp.
ETHICAL CONSIDERATIONS
This research was carried out in order to analyze the antibiotic resistance of the probiotic bacteria found in commercial food products. Probiotics are live microorganisms that confer nutrition and health promoting benefits on the host. Since probiotic bacteria act as a reservoir for antibiotic resistant determinants, they can confer the pathogens’ protection against commonly used drugs. The samples used for this study were curd samples and the antibiotics used to test the antibiotic resistance were tetracycline, erytthromycin, vancomycin and chloramphenicol. The samples were cultured under the appropriate conditions. Bacterial DNA extraction was carried using heat shock method and CTAB method. The DNA concentration was measured. The bacterial isolates were identified using genus specific PCR primers in the conventional PCR. Agarose gel electrophoresis was carried out to visualize the PCR products. Disc diffusion was carried out to detect the antibiotic resistance for the selected antibiotics. This project was solely carried out by myself and my co-author. All the tests were performed at BMS, School of Science, Sri Lanka. All laboratory rules and regulations were abided. Safety precautions were taken at all times when working in the laboratory. All the chemical and biological waste products were disposed appropriately. After the tests, used equipment were washed, cleaned and autoclaved appropriately. In this project no human tissues or clinical products were used. Commercially sensitive data was not revealed. All the data collected was confidentially retained by myself and reserved by my co-author.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.

















